Abstract
Purpose
Crops rely on microbes for critical services, but host benefits can be influenced by local makeup of microbiota and the host’s capacity to select optimal strains. We investigated host benefits that cowpeas receive from microbiota depending on plant genotype, their domestication status, and soil source.
Methods
We performed a full factorial soil inoculation experiment. Twenty diverse cowpea genotypes, selected from wild and domesticated populations, were exposed to soil rinsates from four agricultural sites across California, all having cowpea cultivation and varied physicochemical features. Cowpea investment in and benefit from microbiota was quantified by measuring host growth response to inoculation, nodulation, and segregating trait variation.
Results
Variation in induction of root nodulation and strikingly heterogenous benefits to host growth were observed among soil sites. These effects were restricted to live soil inocula but were absent in autoclaved soil controls that lacked microbiota. Cowpeas expressed heritable variation in nodulation, but there was negligible effect of plant population or domestication status on the net benefit that hosts gained from microbiota.
Conclusion
Soils varied substantially and consistently among cultivation sites and were the most prominent driver shaping host growth effects on cowpeas. While growth benefits vary among host cultivars, soil microbiota (and the conditions that maintain them) predominantly shape plant performance in agricultural settings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant-associated microbial mutualists are abundant, exceptionally diverse, and provide varied services to hosts (Friesen et al. 2011). However, the taxonomic makeup of microbial communities – and consequently the benefits they provide – can vary a great deal over space and time (Heath and Stinchcombe 2014). The drivers that shape soil microbiota can be broadly categorized as top down and bottom up forces. Top down forces are driven by symbiosis traits, host phenotypes that regulate the colonization and infection of associated microbes (Bulgarelli et al. 2013; Porter and Sachs 2020). Symbiosis traits are predicted to play a significant role in shaping symbiont communities (Foster et al. 2017). For instance, plants can release specific flavonoids and other compounds from roots to attract and regulate the growth of microbial partners (Sasse et al. 2018; van Dam and Bouwmeester 2016). Plant exudates can reshape the associated microbial community by enriching or reducing specific microbial taxa on plant roots and in the rhizosphere, and parallel processes occur on leaves (Balachandar et al. 2006; Micallef et al. 2009; Morella et al. 2020). Plants are also thought to impose selection by restricting infection to a subset of microbial strains, and by selectively rewarding or punishing strains post-infection depending on the benefits that they provide (Denison 2000; West et al. 2002). However, symbiosis traits can vary substantially among plant species and even among host genotypes or populations of the same species (Haney et al. 2015; Pahua et al. 2018; Torres-Martínez et al. 2021; Wendlandt et al. 2019), potentially mitigating microbial benefits on plant health, yield, and fitness (Lareen et al. 2016; Mueller and Sachs 2015).
Bottom up forces shape the community makeup of microbes during free-living phases in soil, including abiotic factors such as soil pH, particle size, water availability, nutrient composition, and biotic factors such as microbial predators, competitors, and facilitators (Agler et al. 2016; Bonkowski 2004; Fitzpatrick et al. 2019; Hussain et al. 2018; Leite et al. 2017; Li et al. 2019). In natural settings, soil texture and nutrient availability are primary factors that alter the composition and abundance of bacterial communities (Xu et al. 2018). In managed settings, tillage and fertilization impact soil abiotic factors, affecting species richness and evenness in soil microbial communities (He et al. 2007; Legrand et al. 2018; Zhong et al. 2010). These environmental factors can interact with host selection to drive variation in plant-associated microbial communities (G x E interactions; Peiffer et al. 2013; Wagner et al. 2016). Additionally, the expression of genetic variation for symbiosis traits among related host genotypes can vary with environmental inputs (Batstone et al. 2020; Wood and Brodie 2016). Moreover, symbiosis traits can be degraded in agricultural settings, as domesticated plants often gain less fitness benefits from microbiota than their wild relatives (Porter and Sachs 2020). Staple crops with evidence of reduced benefits from microbiota include soybean, maize, potatoes, wheat, and rice (Bouffaud et al. 2012; Engelhard et al. 2000; Hetrick et al. 2011; Kiers et al. 2007; Zhu et al. 2001). Degradation of symbiosis can be due to artificial selection of above-ground plant traits that tradeoff with belowground symbiosis functions, relaxed selection on belowground traits in rich agricultural settings, or demographic changes in crop plants such as inbreeding or founder effects (Denison 2015; Porter and Sachs 2020). A key aspect of domestication is the movement of plant genotypes to new regions (Gaut et al. 2018), introducing plants to novel soil characteristics and belowground communities which can directly impact host benefits from symbiosis and the expression of symbiosis traits. Examining the relative effects and interplay between host-mediated and environmental forces on soil microbiota and the expression of host symbiosis traits is critical to predicting soil health and plant fitness in natural and agronomic settings.
Plants in the legume family (Fabaceae) associate with rhizobia, proteobacteria that trigger formation of symbiotic root nodules and fix nitrogen (Kakraliya et al. 2018; Sawada et al. 2003), and other rhizosphere associated bacteria that can provide metabolite solubilization, phytostimulation, and other services (Rascovan et al. 2016). Rhizobia can provide substantial amounts of fixed nitrogen, such that host plants can thrive with little or no added nitrogen in the soil (Regus et al. 2017). Individual rhizobia strains, both in natural and agricultural soils, vary tremendously in their effects on hosts, ranging from highly beneficial strains to ones that are ineffective for nitrogen fixation (Gano-Cohen et al. 2020; Moawad and Beck 1991; Thrall et al. 2000). Legumes exert host control by selecting genetically compatible rhizobia and by sanctioning less beneficial strains (Kiers et al. 2003; Heath and Tiffin 2009; Oono et al. 2011; Sachs et al. 2010a, b). These symbiosis traits can vary among legume populations (Heath and Tiffin 2009; Wendlandt et al. 2019). Furthermore, variation in expression of symbiosis traits (such as nodulation) among legume genotypes can be influenced by environmental factors, such as planting location and light availability (Batstone et al. 2020; Heath et al. 2020).
Cowpea (Vigna unguiculata) is a genetically diverse legume with cultivars that require minimal nutrient inputs, offer a high proportion of edible plant mass, and are ideal for regions with limited economic or agricultural resources (Herniter et al. 2020; Muñoz-Amatriaín et al. 2017). Wild cowpeas (subsp. dekindtiana) are distributed throughout Africa and are the progenitors of cultivated cowpea varieties (Ali et al. 2015; Coulibaly et al. 2002). Early domesticated cowpeas, known as landraces, are comprised of two distinct populations, Genepool-1 and Genepool-2 (FST = 0.18), distributed across separate regions in northern and southern Africa, respectively, and each of which is diverged from wild cowpeas (FST = 0.13; Ortiz-Barbosa et al. 2022). The patterns suggest that divergent subsets of wild cowpeas were transported and bred in northern and southern regions of Africa during waves of human migration, with only modest gene flow between them, indicating separate domestication events (Huynh et al. 2013; Muñoz-Amatriaín et al. 2017). Both populations of landraces share a suite of improved traits, including large seeds, shatter-resistant pods, and flexible flowering time (Lo et al. 2018; Xiong et al. 2016). Cowpea landraces are grown under simple agricultural conditions and have not been expanded or adapted to new regions, consistent with an early stage of crop domestication (i.e., stage two of four proposed stages; Gaut et al. 2018).
Here, we examined the roles of plant genotype and soil source in shaping the expression of cowpea host symbiosis traits in response to soil microbiota. We conducted a full factorial soil inoculation experiment where the effects of cowpea host population, genotype, and soil inoculum source were simultaneously analyzed. We used eight wild cowpea genotypes and twelve early-domesticated landraces to examine the role of host genotype – and effects of host domestication – on the expression of host performance and symbiosis traits. We selected cowpea from different populations to account for known variation in symbiosis traits among legume populations and to examine the effects of separate domestication events among the two landrace populations (Ortiz-Barbosa et al. 2022). Plants were inoculated with soil rinsates generated from four agricultural field sites distributed across a 460 km transect in California, having current or recent cultivation with cowpea. We quantified aboveground plant biomass and root-nodulation patterns to estimate host growth response to inoculation. Additionally, we tested whether the differences in the soil sources could influence the expression of segregating variation in symbiosis traits by quantifying soil physicochemical properties and estimating additive genetic variances and heritability among cowpea genotypes. The goals were to i) evaluate the roles of cowpea host genotype and soil source in mediating the expression of plant symbiosis traits, ii) examine whether domestication has influenced plant investment into and benefits from symbiosis when exposed to diverse soil sources, and iii) quantify heritable variation in symbiosis traits and test whether association with diverse soil microbial sources can shape this expression.
Materials and methods
Cowpea genotypes
Eight wild cowpea accessions (i.e., genotypes) were sampled from natural populations in Botswana (PI 632890), Zimbabwe (PI 632891), Tanzania (PI 632876, PI 632892), and Niger (PI 632882, PI 632879, PI 632880, PI 632881). Twelve cowpea landraces were selected from populations in northern and southern Africa. For the northern population, genotypes were sampled from Egypt (TVu-9492), Senegal (TVu-14346), Benin (TVu-8834), Niger (TVu-15591, TVu-14971), and Nigeria (TVu-3804), and southern population genotypes were sampled from Mozambique (NamuesseD, Nhacoongo-3, Muinana-Lawe), Tanzania (TVu-1280), Malawi (TVu-9848), and Zambia (TVu-13305) (Huynh et al. 2013). The African cowpea genotypes are photoperiod sensitive and do not flower or set seed under the summer conditions tested herein. Under shorter day lengths, these lines take about 40 days to flower and 70 days to form pods. Landraces were chosen to maximize genetic diversity and were only selected from germplasm collections made before 1975, after which African breeding programs began transferring cowpea germplasm, leading to admixture among genotypes (Huynh et al. 2013; Ortiz-Barbosa et al. 2022). Accessions were previously genotyped using an Ilumina iSelect Consortium array developed for cowpea, which targets more than 50,000 single-nucleotide polymorphisms. (Muñoz-Amatriaín et al. 2017; Ortiz-Barbosa et al. 2022). Seeds were obtained from the USDA germplasm collection (Griffin, GA).
Soil inocula preparation
Soil sampling locations were selected from fields based on history of agricultural management and sampling accessibility, including at the Coachella Valley Agricultural Research Station in Thermal, CA, the University of California Riverside Agricultural Experiment Station, the Kearney Agricultural Research and Extension Center in Parlier, and a commercial cowpea grower’s field near Shafter, CA (Fig. 1, Table S1). The Thermal, Riverside, and Parlier sites were fallow during sampling and had not been recently irrigated or fertilized, though they did receive low levels of fertilization during prior growth seasons. The grower’s field in Shafter was unique in that it had growing cowpeas at the time of sampling, was recently fertilized, and cowpea had been inoculated via a peat-based seed-coat inoculant prior to planting (Exceed Peat for Cowpea/Lespedeza/Mung Bean, product #: 2013; Visjon Biologics). The conditions of fertilization and seed inoculation at the Shafter site are typical of the current cowpea agricultural process in California (Long et al. 2010). Parlier & Shafter sites were sampled on 6/17/19, Thermal was sampled on 6/21/19, and Riverside was sampled on 6/29/19.
Sampling sites for soils, including a principal components analysis of soil nutrient composition and texture at each site. The x-axis indicates PC1, which explained 45.8% of the soil variation. The y-axis indicates PC2, which explained 33.9% of the variation. Site names, collection dates, sampling coordinates, and crop history from each of the four sites are listed in Table S1
Approximately six liters of topsoil were sampled from four randomized sampling plots at each field site. Soil samples were pooled by field site, sieved, mixed with an equal portion of sterile water, filtered through cheesecloth, left to settle overnight, and the supernatant from each flask was removed (i.e., top ~ 50%) and divided into five portions. This protocol enables plants to be inoculated with dominant microbiota, while minimizing addition of nutrients that could change the soil makeup (Unkovich and Pate 1998). Three portions were reserved at room temperature to be used as a ‘live’ inoculum, while the rest were autoclaved and allowed to cool to serve as a dead control. The next day, seedlings were inoculated with 10 ml of the appropriate inoculum. Live and dead inocula from each site were separately spread inoculated (100 μl) onto plates with a modified arabinose gluconate medium (MAG; Sachs et al. 2009) and incubated at 29 °C for eight days to confirm the presence of soil microbiota in live inocula and likewise confirm the sterility of dead inocula. Live inocula from all four source soils formed dense lawns on the MAG plates, whereas control dead inocula did not generate any colonies. Soil inocula were prepared at two time points from the same sampled soils to account for variation in germination speed among the diverse cowpea genotypes (7/6/19, 8/3/19).
Soil analysis
Soil samples collected at each site in February 2021 were analyzed for organic matter, nitrogen, phosphorus (weak Bray and sodium bicarbonate-P), pH, extractable cations (potassium, magnesium, calcium, sodium), hydrogen, sulfate-S, cation exchange capacity, percent cation saturation, and soil texture (A&L Western Labs, Modesto, CA). A portion of the original soils from 2019 were also analyzed for nitrate nitrogen as a comparison. Principal components analysis (PCA) of quantitative soil measures was performed to reduce dimensionality. Data on soil composition, available water storage, drainage, and proportion of hydric soils were extracted via geolocation from the UC Davis California Soil Research Lab.
Pot and seed preparation
One-gallon nursery pots were filled with wetted soil and autoclaved twice (50:50 silica sand mix of #12 and #30 size). Seeds were surface sterilized in a 6% sodium hypochlorite solution and vortexed intermittently for 3 min, then rinsed four to six times with sterile water, nick-scarified, and planted the same day. Wild cowpea genotypes were planted on 6/12/19 and landraces were planted on 6/19/19 to account for germination timing and growth. Seeds were planted in triplicate per pot and extra seedlings were later removed or redistributed to pots lacking visible growth. Each treatment by genotype combination had 5 live inoculation replicates and 3 controls that received the dead inoculum. These replicates were divided across 8 blocks in the greenhouse, each containing a random arrangement of all treatment combinations (20 plant genotypes × 4 soil sources = 80 plants per block). Controls for each treatment combination were randomly assigned among the eight blocks, with each block containing a mix of live and control-inoculated plants to reduce confounding block effects. Beginning the first week of July, plants with true leaves were fertilized twice weekly with 10 mL of sterilized Jensen’s solution, which contains micronutrients and was supplemented with a minimal concentration of nitrogen to allow for cowpea survival under symbiont free conditions (0.4 g/L of KNO3; Somasegaran and Hoben 1994). Germination was unexpectedly slow for the wild cowpeas, and five additional seeds were planted in pots without visible seedlings on 6/25/19. Prior to inoculation, pots with visible seedlings were rearranged with unsuccessful pots from blocks 1–3 to complete as many blocks as possible. Inoculation of germinated plants (including all landraces and roughly half of the wild plants) took place on 7/7/19. By 7/15/19, nearly all previously planted wild seeds had germinated. These late-germinated plants were then inoculated on 8/4/19. Plants with true leaves were treated with 10 ml of inoculum, directly onto the soil. The greenhouse received weekly pesticide treatments.
Measurement of plant and symbiosis traits
Harvest of plants occurred block by block starting on 8/19/19 and ending on 10/26/19 (Table S2) to account for time necessary to dissect and process plants. Plants which had germinated earlier and received the first round of inoculation were harvested first to minimize variation in growth period. Plants were de-potted, true leaves were counted, and roots were rinsed of soil. Nodules were dissected, counted, photographed, and dried in an oven at 60 °C to weigh biomass. If available, up to ten nodules per plant were set aside prior to drying, surface sterilized, and stored at -80 °C for a separate genotyping study. Roots and shoots were separated and dried in an oven at 60 °C to weigh biomass. To account for the weight of nodules set aside for culturing, fifty nodules of varied size (i.e., nodule radius) were photographed, dried, and weighed individually to generate an area-by-weight curve: estimated nodule mass = 0.00602 + (0.000135 × nodule volume). This curve was used to estimate nodule biomass for plants with 20 or fewer total nodules to reduce potential bias from extrapolation of biomass from low nodule counts. For plants with greater than 20 total nodules, total nodule biomass was estimated by extrapolating from the initial biomass to account for nodules that were set aside for genotyping and were not weighed.
Traits were quantified, including the number of nodules formed, total nodule biomass, mean individual biomass of nodules, total plant biomass, and host growth response. Host growth response was calculated by dividing the total dry biomass of each inoculated plant by the mean dry biomass of the dead inoculum controls of the same genotype. The resulting ratio reflects the effects of inoculated microbiota on plant growth, separate from growth effects due to other soil features (i.e., nutrient variation). This calculation also controls for variation in plant size among cultivars, indicating that genotype or population-level effects in our models are due to variation in response to inoculation, rather than natural size differences (Sachs et al. 2010a, b; Regus et al. 2015; Ortiz-Barbosa et al. 2022).
Linear mixed models were implemented to test whether the trait response varied among soil treatments, among wild and domesticated cowpea populations, and whether differences between wild and domesticated populations depended on the soil treatment while accounting for the cowpea genotypic effects. Soil treatment, cowpea population, and their interaction were treated as fixed factors, and cowpea genotype as a random factor. Days post inoculation was added as a covariate to account for the variation attributed to the different harvest time points. Models with block as a random factor indicated that block was not significant, so it was excluded. For all analyses, host growth and mean nodule biomass were log-transformed, and the number of nodules was square root transformed to meet the assumptions of normality and heteroscedasticity. Tukey’s post-hoc tests were conducted to test for differences among soil treatments and cowpea populations. A variance partitioning test, which assesses the proportionate variation explained by two or more variables, was performed to compare the relative influence of host genotype and soil treatment on host growth response using the publicly-available POV Engine JSL script for JMP, developed by Thomas A. Little Consulting (TLC), 2022. All analyses were performed in JMP® Pro, Version 15.0.0. SAS Institute Inc., Cary, NC, 1989–2022.
Expression of trait genetic variation and heritability
Genetic variation was assessed for symbiosis traits by examining the significance of the random factor with a log-likelihood ratio test between a null model that excluded the genotypic factor and the main model described above. We tested whether the genetic variance component varied significantly among soil inoculum treatments by comparing models with different variance–covariance structures (Shaw 1991; Saxton 2004; Torres-Martínez et al. 2019). A model where the genotype variance component was allowed to vary among soil treatments (heterogeneous variance model) was compared to a model where the genotype variance component was constrained to be identical across soil treatments (homogeneous variance model; Table S3). To evaluate whether a genotype-by-environment (G x E) interaction was observed, we also compared a model where no G x E is assumed with a model where G x E is present (Table S3).
Broad and narrow sense heritability were estimated for traits where a significant genotypic variation was observed. A soil treatment-specific heritability was estimated when the expression of trait genetic variation varied among soil treatments. To better visualize changes in genetic variance, we estimated breeding values of each cowpea genotype under each soil inoculum with Best Linear Unbiased Predictions (BLUPS; Henderson 1975; Liu et al. 2008). BLUPs were calculated from the model that best fit the variation for each trait (Tables S3 and S4). Genetic variation estimates were calculated using the R package sommer (Covarrubias-Pazaran 2016).
Results
Soil physicochemical features
The four soil sources varied in physical and chemical features. Soil textural analysis revealed that all soils were predominantly sandy (i.e., particles 50–2000 μm in diameter), but that the Thermal soil had the highest sand proportion (78%, compared to an average of 54% for the remaining sites; Table S5). The first principal component (PC1) of the quantitative soil analysis explained 45.8% of the variation in physicochemical properties (Fig. 1, Table S5). PC1 was mainly driven by variation in the proportion of silt and sand particles, available phosphorous, and salinity (Table S5).
Parlier and Riverside field sites were classified by the Hanford soil series, with Shafter and Thermal sites classified by the Lewkalb & Myoma series, respectively (Table S1). Both Hanford and Lewkalb soils are characterized as deep, well-drained, coarse-loamy, and mixed, while Lewkalb soils are also calcareous (Soil Survey Staff, USDA). Myoma soils are characterized by fine, moderately alkaline sands which are somewhat excessively drained (Soil Survey Staff, USDA).
Symbiosis trait variation
Total plant biomass
Live and dead soil treatments resulted in significant differences in plant biomass, as did the soil source and the interaction effect (live/dead x soil source; Table S6). Cowpea host population and genotype also had significant effects on plant biomass, indicating natural size variation among genotypes not due to inoculation. In all models for total plant biomass, days post inoculation was a significant factor and was therefore included as a covariate in analysis (Tables S6 & S7).
Host growth response
Live soil treatments had significant positive effects on host growth, with significant variation among soil sources that ranged 2-3X in magnitude (Fig. 2a). In contrast, treatment with sterilized dead soils did not produce any significant differences in total plant biomass among soil treatments (Table S7). While total plant biomass did vary significantly among cowpea populations (Table S7), indicating natural differences in plant size, host growth response to inoculation did not vary significantly among cowpea populations, and mean host growth response values by population did not vary by soil treatment (no Population x Treatment interaction effect, Table 1). Host genotype had a significant effect on host growth response, but these differences were modest, and most genotypes (16/20) were not significantly different from one another (Table 1; Fig. 3). A partition of variance (POV) test indicated that differences among soil treatments explained almost twice the variance in host growth response (25.98%) compared to differences among host genotypes (14.94%; Table 2). While host genotype and soil source were both significant factors in our model of host growth response, these data indicate that variation in soil treatment (rather than host domestication or provenance) was the prominent factor mediating host benefits. Days post inoculation was also a significant factor and was included as a covariate during analysis (Table 1).
Variation in symbiosis traits among populations. Boxplots of (a) Host growth response, (b) Number of nodules, (c) Total nodule biomass, and (d) Mean nodule biomass in response to inoculation from 4 distinct sites and across three populations of African cowpea (two landrace populations and one wild population). Treatments are denoted by color (pink = Parlier, green = Riverside, blue = Shafter, purple = Thermal). Connected letters represent Tukey groupings from linear mixed models, calculated within each lineage. For both Total nodule biomass and Mean nodule biomass, plants without nodules were excluded. Host growth and mean nodule biomass were log-transformed, and the number of nodules was squared root transformed. Outliers are hidden for visual simplicity
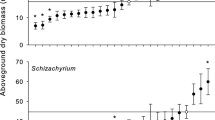
Variation in host growth response (HGR) among host genotypes. Grouped interval plot indicates standard deviation in response to inoculation from soil treatments, denoted by color (pink = Parlier, green = Riverside, blue = Shafter, purple = Thermal) across twenty cowpea lines, ordered from highest to lowest mean HGR. Connected letters show Tukey groupings. Global mean HGR and standard deviation of each host genotype is shown in black. Boxes below each host genotype denote the host population (white = wild, grey = landrace population 1, black = landrace population 2), showing that host populations are highly intermixed when genotypes are ranked by HGR
Number of nodules
Host population and soil source both had significant effects on the number of nodules formed, with a significant interaction effect (Table 1). The wild cowpea population formed significantly fewer nodules (mean = 35.5 ± 2.8) than either landrace populations 1 or 2 (86.3 ± 8.2; 93.6 ± 7.8, respectively; Table S8) and there were also significant differences between the landrace populations (Table S9). When including total plant biomass as a covariate, the wild population still formed significantly fewer nodules than either landrace population 1 or 2, respectively (T367 = 3.14, p < 0.01; T367 = 3.87, p < 0.01). Days post inoculation also had a significant effect on nodulation and was included as a covariate during analysis (Table 1).
Each soil treatment resulted in significantly different nodule counts (Table S9). Despite the similar appearance of treatment ranking among populations with regard to nodulation (Fig. 2b), there was a significant Population x Treatment effect (Table 1). The Shafter soil inoculation induced nodules in only 59 of 98 plants (~ 60%). In contrast, the Thermal soil induced nodulation in 98% of plants, and Parlier and Riverside soils had 100% nodulation. Within the Parlier and Riverside inoculum treatment groups, both landrace populations formed significantly more nodules than the wild population, while within the Thermal and Shafter treatment groups, landrace population 2 formed significantly more nodules than either of the other populations (Table S9).
Twelve of 239 control plants had nodules (~ 5%), indicating contamination, and were excluded from analysis (Table S2). Nine of the contaminated plants had 8 or fewer nodules, whereas the mean nodule count for an inoculated plant was 66. We were unable to detect potential cross-contamination by other microbiota. Additionally, 28 plants had lost over 50% of their leaves, indicating senescence likely due to stress from late-harvest pest control spray treatments, or had mature seed pods, indicating senescence due to shorter day lengths. These plants were also excluded from analysis. Among these senesced plants, 16 individuals (64%) belonged to two host genotypes, TVu-1280 and TVu-9848, both from landrace population 2. One individual was incorrectly harvested at 22 days post inoculation. For all remaining plants, days post inoculation ranged from 42 to 105 days. The majority of plants were harvested within two weeks of the mean days post inoculation (66 days, 52% of plants).
Nodule biomass
Host population, soil source, and their interaction all had significant effects on total and mean individual nodule biomass (Table 1). Nodules from landrace population 1 were the largest, and population 2 were the smallest nodules (Tables 2 and 3). Shafter and Thermal soils induced significantly larger nodules on average than either of the other treatment groups, despite their association with lower levels of host growth (Table S9, Fig. 2d).
Both cowpea landrace populations had a higher total nodule biomass than the wild population (49% and 77% higher, respectively), which was consistent across most treatments (Table S9). We found no significant differences in total nodule biomass among soil treatments within landrace population 2 or the wild population (Fig. 2c). With total plant biomass as a covariate, the wild population still had a lower total nodule biomass than either landrace 1 or 2 populations (T325 = 3.07, p < 0.01; T325 = 5.06, p < 0.01). These data suggest that wild cowpeas had a proportionally lower investment into nodule tissues. Soil from Thermal induced the highest total nodule biomass (mean = 202.72 mg, Table S8), which was significantly higher than both the Shafter and Riverside soils. Days post inoculation had a significant effect on total nodule biomass and was included as a covariate; however, days post inoculation was not a significant factor in our model for mean nodule biomass.
Effects of soil characteristics on symbiosis traits
In a linear mixed model with PC1 (from the quantitative soil analysis), host population, and their interaction as fixed effects and host genotype as a random effect, we found that PC1 had a significant effect on host growth response, number of nodules, and mean nodule biomass (Table 3). We also found a significant host population x PC1 interaction effect for both number of nodules and mean nodule biomass (Table 3). Conversely, we found no significant differences among autoclaved inoculum treatments. These data suggest that growth differences among soil treatments are driven primarily by variation in microbial community, which is modulated by the soil physicochemical characteristics (Table S7). For all traits except for number of nodules, days post inoculation was a significant factor, and was included as a covariate (Table 3).
Cowpea genetic variation and heritability
The expression of genetic variation (σ2G) for the number of nodules and total nodule biomass varied with the soil inoculum imposed, respectively (χ2 5 = 21.85, p < 0.01; χ2 5 = 14.20, p = 0.01), but host growth response and mean nodule biomass did not (χ2 5 = 2.3, p = 0.80; χ2 5 = 10.6, p = 0.06; Fig. 4; Table S3), consistent with soil rather than plant genotype being the prominent driver shaping host growth effects on cowpeas, despite both host genotype and soil affecting nodulation patterns. The highest expression of σ2G for the number of nodules was observed within the soil inoculum from Parlier, and the lowest σ2G was observed within the soil inoculum from Thermal, further indicating the highest and lowest heritability for this trait, respectively (Table S4). These patterns were maintained for the additive genetic variation (σ2A) when considering the additive relationship among cowpea genotypes (Table S4). With total nodule biomass, the highest expression of σ2G and heritability was observed within Shafter followed by Thermal, and no genetic variation was evident within Riverside and Parlier, which shared the Hanford soil composition (Table S4). The addition of the relationship matrix caused an overfit of the model and estimates of σ2A within each soil treatment were not obtained for total nodule biomass, so narrow sense heritability was excluded from total nodule biomass reports (Table S4).
Reaction norms of symbiosis traits with a significant genotype effect. (a) Host growth response, (b) Number of nodules, (c) Total nodule biomass, (d) Mean nodule biomass. In the y-axis are the estimated breeding values for each genotype based on adjusted BLUP values from each variance–covariance model that best fit the data. These values were back transformed to their original scale. Each dot represents an individual cowpea genotype
A G x E interaction was observed for both host growth and mean nodule biomass (χ2 1 = 6.11, p = 0.01; χ2 1 = 7.97, p < 0.01; Table S3) despite the homogeneity in genetic variances among soil treatments for these traits (Fig. 4). For the number of nodules and total nodule biomass, a significant G x E was evident when genetic variances and covariances were allowed to differ among treatments, indicating differences in the phenotypic plasticity of cowpea genotypes, as well as genetic variation in phenotypic plasticity (Table S3).
Discussion
We found that soil source strongly influenced both host benefits and expression of belowground plant traits in the cowpea-rhizobia symbiosis. Soil source was a significant factor contributing to host growth response and nodule counts (Fig. 2, Table 1), and soil composition appeared to play a prominent role in these effects. Both Riverside and Parlier sites – which induced the strongest host growth response – share the same soil series type (Hanford), and a similar among-genotype variation was observed in these soil sources for both nodule counts and total nodule biomass (Tables S1, S4). Conversely, the Shafter and Thermal soils have distinct soil compositions (Lewkalb, Myoma) and different among-genotype variation for these same traits (Tables S1, S4). We also found that soil physicochemical properties (PC1) had a significant effect on host growth response, number of nodules, and mean nodule biomass (Table 3). As there were no significant differences in plant biomass among the dead inoculum controls due to either soil treatment or PC1 (Table S7), this suggests that soil physicochemical properties shape microbiota in each soil, thus indirectly driving plant benefits from inoculation. Our analysis of trait heritability suggests that different soil treatments can shift the expression of genetic variation in the number and size of nodules, but not for the host growth response of cowpea. For host growth, we found a significant G x E interaction, suggesting the presence of differences in phenotypic plasticity of host growth in cowpea genotypes in response to the soil rhizobia community.
Previous studies have also suggested soil-driven effects in the cowpea-microbial symbiosis. For instance, soil particle makeup and pH influenced the rhizobia populations in cowpeas sampled from agronomic fields in Kenya, as well as rhizobia cultured from nearby uncultivated soils (Ndungu et al. 2018). Similarly, soil type played a larger role than plant genotype in shaping non-rhizobia cowpea nodule microbial communities (Leite et al. 2017). However, neither of these studies examined the effect of soil conditions on plant growth or benefits from those microbes. Other studies that focus on legume inoculation benefits have shown that host genotype, inoculation, and soil type are all significant drivers of host growth and nodulation; however, in each case, plants were inoculated with a single strain of rhizobia (Amha and Fassil 2018; Keller and Lau 2018; Sánchez et al. 2014).
It was striking that no significant differences in host benefits from soil inoculation were observed among cowpea populations, given that the cowpea genotypes span the diversity of this species (Huynh et al. 2013). This also supports the hypothesis that domestication has not degraded cowpea symbiosis benefits, as wild and domesticated cowpea respond similarly when treated with the same soil communities (Ortiz-Barbosa et al. 2022). Nonetheless, landrace population 2 formed significantly more nodules than population 1 with the soil treatments from Thermal and Shafter, and had a significantly higher host growth response than population 1 within the Thermal treatment (Table S9). This indicates that landrace population 2 might be more resilient under challenging soil conditions, as the Thermal and Shafter treatment groups were the least beneficial overall (Table S8, Fig. 2). However, there are also limitations in our approach that should be considered. Preparation of soil for inoculation can change qualitative and functional diversity of rhizobia present (Alberton et al. 2006). Additionally, some of the observed soil inoculation effects could be due to density, rather than community makeup, of compatible microbes that varied among sites. In particular, low nodulation effects from the Shafter inoculum could indicate either a reduced or significantly altered rhizobial population. Nonetheless, for growers considering different cowpea cultivars as well as different field plots, our data suggests that the field soil – and the microbial community it contains – is more important for determining yield. Additionally, analysis of genotypic variation & expression among specific genotypes suggests that cowpea genotypes respond to changes in soil microbial communities in different ways, and that a change in soil inoculum can alter the ranking of genotypes when examining host growth (Table S10). This is a factor which should be considered by plant breeders and those making planting decisions, when cultivar-specific consistency in growth response to inocula is desirable.
The lower nodulation and host growth associated with the Shafter soil inoculation was surprising, as this was the only soil that had been treated with a Bradyrhizobium biofertilizer, as well as the only inoculum from a field with live cowpea at the time of sampling, both factors that we expected to enhance nodulation. However multiple factors can mediate the success of inoculation. When inocula were derived from field soils which had been recently fertilized, treated plants experienced significantly reduced biomass compared with non-fertilized soil inoculation, suggesting that fertilization impacts soil populations of nitrogen-fixing rhizobia (Simonsen et al. 2015). Long term field nitrogen fertilization has also been shown to stimulate the evolution of less-mutualistic rhizobia strains (Klinger et al. 2016; Weese et al. 2015). However, chemical analysis of Shafter soils showed that in 2019, the nitrogen levels (NO3-N) at this site were low relative to other sites (Table S5), suggesting that any negative impacts to local microbiota are likely not due to recent fertilization. The crop history at Shafter might also explain some of the variation seen, as each of the other soils originated from sites where a multi-parent intercross population of cowpea genotypes have been propagated for multiple seasons (Huynh et al. 2018), while Shafter had a mixed crop history. In the 3 years prior to sampling, the Shafter field had been used to grow carrots and cotton. Prior to that, it had been an alfalfa field for 4 years. Like cowpea, alfalfa is a legume; however, it generally associates with Ensifer, and does not form nodules with Bradyrhizobium (Stajković-Srbinović et al. 2012; Wang et al. 2018; Woliy et al. 2019). Bioinoculants such as the one used at Shafter are employed to prime soils without a history of successful prior production of a particular legume, making this site a perfect candidate for inoculation. Since Shafter was sampled during the first season of cowpea growth, it’s possible that the soils had not yet been sufficiently enriched with Bradyrhizobium.
Later stages of plant domestication often involve the introduction of plant genotypes to new regions and thus to soils with novel characteristics and microbial communities (Gaut et al. 2018). Thus, while the African cowpea populations might not be adapted to microbes in Californian soils, introduction to novel soils and microbes is a fundamental aspect of agriculture (Gaut et al. 2018). We found that the expression of genetic variation in host growth response to soil treatments did not vary significantly among cowpea genotypes, and that variance in host growth response was more strongly associated with soil treatment than host genotype, suggesting that field soil locations (and their associated microbiota) are more important than host cultivar when predicting host benefits and expected yields. The yield gap – the difference between actual and maximum expected crop yield – is substantial for cowpeas grown in Africa (Foyer et al. 2016). Like other domesticated legumes, modern cowpeas are most often fertilized to maximize growth, suggesting that key below-ground traits have been lost or neglected in the process of domestication (Denison 2000). However, we did not find significant differences in host growth response among wild and landrace populations, confirming results from Ortiz-Barbosa et al. (2022) that early cowpea domestication has not degraded host benefits from symbiosis. Future studies could illuminate how rhizobia communities in nodules vary among wild and domesticated genotypes. With the increase in above-ground plant mass associated with domestication, cowpea could have adapted strategies to maintain fitness benefits. If so, identifying these traits would prove useful in breeding cowpea and other domesticated legumes to harness local rhizobia, improve crop yields, and reduce inorganic fertilization practices.
Data availability
Data supporting the findings of this study are available in the supplementary material of this article.
References
Agler MT, Ruhe J, Kroll S et al (2016) Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol 14:e1002352. https://doi.org/10.1371/journal.pbio.1002352
Alberton O, Kaschuk G, Hungria M (2006) Sampling effects on the assessment of genetic diversity of rhizobia associated with soybean and common bean. Soil Biol Biochem 38:1298–1307. https://doi.org/10.1016/j.soilbio.2005.08.018
Ali ZB, Yao KN, Odeny DA et al (2015) Assessing the genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] accessions from Sudan using simple sequence repeat (SSR) markers. Afr J Plant Sci 9:293–304. https://doi.org/10.5897/AJPS2015.1313
Amha G, Fassil A (2018) The effect of inter cross-inoculation host group rhizobia on the growth and nitrogen fixation of Faba Bean (Vicia faba L.) varieties in North Showa, Amhara Regional State, Ethiopia. J Agric Biotech Sustain Dev 10:25–33. https://doi.org/10.5897/JABSD2018.0307
Balachandar D, Sandhiya GS, Sugitha TCK, Kumar K (2006) Flavonoids and Growth Hormones Influence Endophytic Colonization and in Planta Nitrogen Fixation by a Diazotrophic Serratia sp. in Rice. World J Microbiol Biotechnol 22:707–712. https://doi.org/10.1007/s11274-005-9094-0
Batstone RT, Peters MAE, Simonsen AK et al (2020) Environmental variation impacts trait expression and selection in the legume–rhizobium symbiosis. Am J Bot 107:195–208. https://doi.org/10.1002/ajb2.1432
Bonkowski M (2004) Protozoa and plant growth: the microbial loop in soil revisited. New Phytol 162:617–631. https://doi.org/10.1111/j.1469-8137.2004.01066.x
Bouffaud M-L, Kyselková M, Gouesnard B et al (2012) Is diversification history of maize influencing selection of soil bacteria by roots? Mol Ecol 21:195–206. https://doi.org/10.1111/j.1365-294X.2011.05359.x
Bulgarelli D, Schlaeppi K, Spaepen S et al (2013) Structure and Functions of the Bacterial Microbiota of Plants. Annu Rev Plant Biol 64:807–838. https://doi.org/10.1146/annurev-arplant-050312-120106
Coulibaly S, Pasquet RS, Papa R, Gepts P (2002) AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor Appl Genet 104:358–366. https://doi.org/10.1007/s001220100740
Covarrubias-Pazaran G (2016) Genome-Assisted Prediction of Quantitative Traits Using the R Package sommer. PLoS ONE 11:e0156744. https://doi.org/10.1371/journal.pone.0156744
Denison RF (2000) Legume Sanctions and the Evolution of Symbiotic Cooperation by Rhizobia. Am Nat 156:567–576. https://doi.org/10.1086/316994
Denison RF (2015) Chapter 9 - A Darwinian perspective on improving nitrogen-fixation efficiency of legume crops and forages. In: Sadras VO, Calderini DF (eds) Crop Physiology, 2nd edn. Academic Press, San Diego, pp 207–222
Engelhard M, Hurek T, Reinhold-Hurek B (2000) Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ Microbiol 2:131–141. https://doi.org/10.1046/j.1462-2920.2000.00078.x
Fitzpatrick CR, Mustafa Z, Viliunas J (2019) Soil microbes alter plant fitness under competition and drought. J Evol Biol 32:438–450. https://doi.org/10.1111/jeb.13426
Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S (2017) The evolution of the host microbiome as an ecosystem on a leash. Nature 548:43–51. https://doi.org/10.1038/nature23292
Foyer CH, Lam H-M, Nguyen HT et al (2016) Neglecting legumes has compromised human health and sustainable food production. Nat Plants 2:16112. https://doi.org/10.1038/nplants.2016.112
Friesen ML, Porter SS, Stark SC et al (2011) Microbially Mediated Plant Functional Traits. Annu Rev Ecol Evol Syst 42:23–46. https://doi.org/10.1146/annurev-ecolsys-102710-145039
Gano-Cohen K, Wendlandt C, Moussawi K et al (2020) Recurrent mutualism breakdown events in a legume rhizobia metapopulation. Proc R Soc B Biol Sci 287:20192549. https://doi.org/10.1098/rspb.2019.2549
Gaut BS, Seymour DK, Liu Q, Zhou Y (2018) Demography and its effects on genomic variation in crop domestication. Nat Plants 4:512–520. https://doi.org/10.1038/S41477-018-0210-1
Haney CH, Samuel BS, Bush J, Ausubel FM (2015) Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants 1:1–9. https://doi.org/10.1038/nplants.2015.51
He J-Z, Shen J-P, Zhang L et al (2007) Quantitative Analyses of the Abundance and Composition of Ammonia-Oxidizing Bacteria and Ammonia-Oxidizing Archaea of a Chinese Upland Red Soil under Long-Term Fertilization Practices. Environ Microbiol 9:2364–2374. https://doi.org/10.1111/j.1462-2920.2007.01358.x
Heath KD, Podowski JC, Heniff S et al (2020) Light availability and rhizobium variation interactively mediate the outcomes of legume–rhizobium symbiosis. Am J Bot 107:229–238. https://doi.org/10.1002/ajb2.1435
Heath KD, Stinchcombe JR (2014) Explaining Mutualism Variation: A New Evolutionary Paradox? Evolution 68:309–317. https://doi.org/10.1111/evo.12292
Heath KD, Tiffin P (2009) Stabilizing Mechanisms in a Legume-Rhizobium Mutualism. Evol 63:652–662 https://doi.org/10.1111/j.1558-5646.2008.00582.x
Henderson CR (1975) Best Linear Unbiased Estimation and Prediction under a Selection Model. Biometrics 31:423–447. https://doi.org/10.2307/2529430
Herniter IA, Muñoz-Amatriaín M, Close TJ (2020) Genetic, textual, and archeological evidence of the historical global spread of cowpea (Vigna unguiculata [L.] Walp.). Legume Sci 2:e57 https://doi.org/10.1002/leg3.57
Hetrick B, Wilson G, Cox T (2011) Mycorrhizal dependence of modern wheat varieties, landraces, and ancestors. Can J Bot 70:2032–2040. https://doi.org/10.1139/b92-253
Hussain M, Hamid MI, Tian J, et al. (2018) Bacterial community assemblages in the rhizosphere soil, root endosphere and cyst of soybean cyst nematode-suppressive soil challenged with nematodes. FEMS Microbiol Ecol 94:fiy142 https://doi.org/10.1093/femsec/fiy142
Huynh B-L, Close TJ, Roberts PA, et al. (2013) Gene Pools and the Genetic Architecture of Domesticated Cowpea. Plant Genome 6:plantgenome2013.03.0005 https://doi.org/10.3835/plantgenome2013.03.0005
Huynh B-L, Ehlers JD, Huang BE et al (2018) A multi-parent advanced generation inter-cross (MAGIC) population for genetic analysis and improvement of cowpea (Vigna unguiculata L. Walp.). Plant J 93:1129–1142. https://doi.org/10.1111/tpj.13827
Kakraliya SK, Singh U, Bohra A et al (2018) Nitrogen and Legumes: A Meta-analysis. In: Meena RS, Das A, Yadav GS, Lal R (eds) Legumes for Soil Health and Sustainable Management. Springer, Singapore, pp 277–314
Keller KR, Lau JA (2018) When mutualisms matter: Rhizobia effects on plant communities depend on host plant population and soil nitrogen availability. J Ecol 106:1046–1056. https://doi.org/10.1111/1365-2745.12938
Kiers ET, Hutton MG, Denison RF (2007) Human selection and the relaxation of legume defences against ineffective rhizobia. Proc R Soc B Biol Sci 274:3119–3126. https://doi.org/10.1098/rspb.2007.1187
Kiers ET, Rousseau RA, West SA, Denison RF (2003) Host sanctions and the legume-rhizobium mutualism. Nature 425:78–81. https://doi.org/10.1038/nature01931
Klinger CR, Lau JA, Heath KD (2016) Ecological genomics of mutualism decline in nitrogen-fixing bacteria. Proc R Soc B Biol Sci 283:20152563. https://doi.org/10.1098/rspb.2015.2563
Lareen A, Burton F, Schäfer P (2016) Plant root-microbe communication in shaping root microbiomes. Plant Mol Biol 90:575–587. https://doi.org/10.1007/s11103-015-0417-8
Legrand F, Picot A, Cobo-Díaz JF et al (2018) Effect of tillage and static abiotic soil properties on microbial diversity. Appl Soil Ecol 132:135–145. https://doi.org/10.1016/j.apsoil.2018.08.016
Leite J, Fischer D, Rouws LFM, et al. (2017) Cowpea Nodules Harbor Non-rhizobial Bacterial Communities that Are Shaped by Soil Type Rather than Plant Genotype. Front Plant Sci 7https://doi.org/10.3389/fpls.2016.02064
Li M, Wei Z, Wang J et al (2019) Facilitation promotes invasions in plant-associated microbial communities. Ecol Lett 22:149–158. https://doi.org/10.1111/ele.13177
Liu XQ, Rong JY, Liu XY (2008) Best linear unbiased prediction for linear combinations in general mixed linear models. J Multivar Anal 99:1503–1517. https://doi.org/10.1016/j.jmva.2008.01.004
Lo S, Muñoz-Amatriaín M, Boukar O, et al. (2018) Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci Rep 8:6261 https://doi.org/10.1038/S41598-018-24349-4
Long R, Temple S, Schmierer J et al (2010) Common dry bean production in California, 2nd edn. University of California, Agriculture and Natural Resources. https://doi.org/10.3733/ucanr.8402
Micallef SA, Shiaris MP, Colón-Carmona A (2009) Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot 60:1729–1742. https://doi.org/10.1093/jxb/erp053
Moawad H, Beck DP (1991) Some characteristics of Rhizobium leguminosarum isolates from uninoculated field-grown lentil. Soil Biol Biochem 23:933–937. https://doi.org/10.1016/0038-0717(91)90173-H
Morella NM, Weng FC-H, Joubert PM et al (2020) Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. PNAS 117:1148–1159. https://doi.org/10.1073/pnas.1908600116
Mueller UG, Sachs JL (2015) Engineering Microbiomes to Improve Plant and Animal Health. Trends Microbiol 23:606–617. https://doi.org/10.1016/j.tim.2015.07.009
Muñoz-Amatriaín M, Mirebrahim H, Xu P et al (2017) Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J 89:1042–1054. https://doi.org/10.1111/tpj.13404
Ndungu SM, Messmer MM, Ziegler D et al (2018) Cowpea (Vigna unguiculata L. Walp) hosts several widespread bradyrhizobial root nodule symbionts across contrasting agro-ecological production areas in Kenya. Agric Ecosyst Environ 261:161–171. https://doi.org/10.1016/j.agee.2017.12.014
Oono R, Anderson CG, Denison RF (2011) Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc R Soc B Biol Sci 278:2698–2703. https://doi.org/10.1098/rspb.2010.2193
Ortiz-Barbosa GS, Torres-Martínez L, Manci A, et al. (2022) No disruption of rhizobial symbiosis during early stages of cowpea domestication. Evolution evo.14424. https://doi.org/10.1111/evo.14424
Pahua VJ, Stokes PJN, Hollowell AC et al (2018) Fitness variation among host species and the paradox of ineffective rhizobia. J Evol Biol 31:599–610. https://doi.org/10.1111/jeb.13249
Peiffer JA, Spor A, Koren O et al (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. PNAS 110:6548–6553. https://doi.org/10.1073/pnas.1302837110
Porter SS, Sachs JL (2020) Agriculture and the Disruption of Plant-Microbial Symbiosis. Trends Ecol Evol 35:426–439. https://doi.org/10.1016/j.tree.2020.01.006
Rascovan N, Carbonetto B, Perrig D et al (2016) Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci Rep 6:28084. https://doi.org/10.1038/srep28084
Regus JU, Gano KA, Hollowell AC et al (2015) Lotus hosts delimit the mutualism–parasitism continuum of Bradyrhizobium. J Evol Biol 28:447–456. https://doi.org/10.1111/jeb.12579
Regus JU, Wendlandt CE, Bantay RM et al (2017) Nitrogen deposition decreases the benefits of symbiosis in a native legume. Plant Soil 414:159–170. https://doi.org/10.1007/s11104-016-3114-8
Sachs JL, Kembel SW, Lau AH, Simms EL (2009) In Situ Phylogenetic Structure and Diversity of Wild Bradyrhizobium Communities. Appl Environ Microbiol 75:4727–4735. https://doi.org/10.1128/AEM.00667-09
Sachs JL, Russell JE, Lii YE et al (2010a) Host control over infection and proliferation of a cheater symbiont. J Evol Biol 23:1919–1927. https://doi.org/10.1111/j.1420-9101.2010.02056.x
Sachs JL, Ehinger MO, Simms EL (2010b) Origins of cheating and loss of symbiosis in wild Bradyrhizobium. J Evol Biol 23:1075–1089. https://doi.org/10.1111/j.1420-9101.2010.01980.x
Sánchez AC, Gutiérrez RT, Santana RC et al (2014) Effects of co-inoculation of native Rhizobium and Pseudomonas strains on growth parameters and yield of two contrasting Phaseolus vulgaris L. genotypes under Cuban soil conditions. Eur J Soil Biol 62:105–112. https://doi.org/10.1016/j.ejsobi.2014.03.004
Sasse J, Martinoia E, Northen T (2018) Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci 23:25–41. https://doi.org/10.1016/j.tplants.2017.09.003
Sawada H, Kuykendall LD, Young JM (2003) Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J Gen Appl Microbiol 49:155–179. https://doi.org/10.2323/jgam.49.155
Saxton AM (2004) Genetic analysis of complex traits using SAS. SAS Institute
Shaw RG (1991) The Comparison of Quantitative Genetic Parameters Between Populations. Evolution 45:143–151. https://doi.org/10.1111/j.1558-5646.1991.tb05273.x
Simonsen AK, Han S, Rekret P et al (2015) Short-term fertilizer application alters phenotypic traits of symbiotic nitrogen fixing bacteria. PeerJ 3:e1291. https://doi.org/10.7717/peerj.1291
Somasegaran P, Hoben HJ (1994) Handbook for Rhizobia: Methods in legume-Rhizobium technology. xvi, 450. https://doi.org/10.1007/978-1-4613-8375-8
Stajković-Srbinović O, De Meyer SE, Miličić B et al (2012) Genetic diversity of rhizobia associated with alfalfa in Serbian soils. Biol Fertil Soils 48:531–545. https://doi.org/10.1007/s00374-011-0646-1
Thrall PH, Burdon JJ, Woods MJ (2000) Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian legumes: interactions within and between genera. J Appl Ecol 37:52–65. https://doi.org/10.1046/j.1365-2664.2000.00470.x
Torres-Martínez L, McCarten N, Emery NC (2019) The adaptive potential of plant populations in response to extreme climate events. Ecol Lett 22:866–874. https://doi.org/10.1111/ele.13244
Torres-Martínez L, Porter SS, Wendlandt C et al (2021) Evolution of specialization in a plant-microbial mutualism is explained by the oscillation theory of speciation. Evolution 75:1070–1086. https://doi.org/10.1111/evo.14222
Unkovich MJ, Pate JS (1998) Symbiotic effectiveness and tolerance to early season nitrate in indigenous populations of subterranean clover rhizobia from S.W. Australian pastures. Soil Biol Biochem 30:1435–1443. https://doi.org/10.1016/S0038-0717(97)00258-7
van Dam NM, Bouwmeester HJ (2016) Metabolomics in the Rhizosphere: Tapping into Belowground Chemical Communication. Trends Plant Sci 21:256–265. https://doi.org/10.1016/j.tplants.2016.01.008
Wagner MR, Lundberg DS, del Rio TG et al (2016) Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun 7:12151. https://doi.org/10.1038/ncomms12151
Wang XL, Cui WJ, Feng XY et al (2018) Rhizobia inhabiting nodules and rhizosphere soils of alfalfa: A strong selection of facultative microsymbionts. Soil Biol Biochem 116:340–350. https://doi.org/10.1016/j.soilbio.2017.10.033
Weese DJ, Heath KD, Dentinger BTM, Lau JA (2015) Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution 69:631–642. https://doi.org/10.1111/evo.12594
Wendlandt CE, Regus JU, Gano-Cohen KA et al (2019) Host investment into symbiosis varies among genotypes of the legume Acmispon strigosus, but host sanctions are uniform. New Phytol 221:446–458. https://doi.org/10.1111/nph.15378
West SA, Kiers ET, Simms EL, Denison RF (2002) Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc Biol Sci 269:685–694. https://doi.org/10.1098/rspb.2001.1878
Woliy K, Degefu T, Frostegård Å (2019) Host Range and Symbiotic Effectiveness of N2O Reducing Bradyrhizobium Strains. Front Microbiol 10:2746. https://doi.org/10.3389/fmicb.2019.02746
Wood CW, Brodie ED III (2016) Evolutionary response when selection and genetic variation covary across environments. Ecol Lett 19:1189–1200. https://doi.org/10.1111/ele.12662
Xiong H, Shi A, Mou B, et al. (2016) Genetic Diversity and Population Structure of Cowpea (Vigna unguiculata L. Walp). PLoS One 11:e0160941 https://doi.org/10.1371/journal.pone.0160941
Xu Z, Yu G, Zhang X et al (2018) Biogeographical patterns of soil microbial community as influenced by soil characteristics and climate across Chinese forest biomes. Appl Soil Ecol 124:298–305. https://doi.org/10.1016/j.apsoil.2017.11.019
Zhong W, Gu T, Wang W et al (2010) The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil 326:511–522. https://doi.org/10.1007/s11104-009-9988-y
Zhu Y-G, Smith SE, Barritt AR, Smith FA (2001) Phosphorus (P) efficiencies and mycorrhizal responsiveness of old and modern wheat cultivars. Plant Soil 237:249–255. https://doi.org/10.1023/A:1013343811110
Acknowledgements
We thank Bao Lam Huynh and Timothy J. Close for providing seeds for the cowpea genotypes used in our experiments, and Peter J. N. Stokes for assistance with generating inbred lines.
Funding
This research was funded by a Delfino Agriculture Technology grant, USDA Hatch grant CA-R-EEOB-5200-H, NIFA-USDA 2022–67019-36500, and by the NSF 1738009 (all to JLS).
Author information
Authors and Affiliations
Contributions
Max Manci and Joel L. Sachs designed the research. Max Manci, Oscar G. Mercado, Raphiel X. Camantigue, Fizzah Khairi, Sierra Neal, Warisha F. Farsamin, and Matthew T. Lampe performed the experiment, and Max Manci, Oscar G. Mercado, Raphiel X. Camantigue, Teresa Nguyen, Warisha F. Farsamin, Matthew T. Lampe, Ivan A. Perez, Jacob Rothschild, Tram H. Le, and Gabriel S. Ortiz-Barbosa collected the data. Max Manci, Lorena Torres- Martínez, and Joel L. Sachs analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Responsible Editor: Didier Lesueur.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Manci, M., Mercado, O.G., Camantigue, R.X. et al. Live soil inocula, not host population or domestication status, is the predominant driver of growth benefits to cowpea. Plant Soil 482, 585–600 (2023). https://doi.org/10.1007/s11104-022-05709-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05709-6