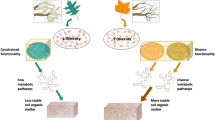
Abstract
Aims
We investigated the role of plants and their plant-derived carbon in shaping the microbial community that decomposes substrates and traced the return of nutrients from decomposition back to plant shoots in order to understand the importance of plants for ecosystem element cycling.
Methods
We performed a greenhouse experiment having plant communities with and without arbuscular mycorrhizal fungi (AMF) and ingrowth cores that held different 15N labeled substrates. We determined the microbial community structure using molecular sequencing and the net assimilation of plant carbon into soil microorganisms using a 13CO2 pulse and 13C measurements of microbial biomarkers. We determined the return of nitrogen back to the shoots using the 15N signal, which was provided from the decomposition of the substrate added to the ingrowth cores.
Results
We observed that the microbial community composition in the ingrowth cores and their net 13C assimilation depended on the presence of AMF and the added substrate. Both plant communities had similar 15N uptake into their shoots, but the net N uptake cost was significantly lower in presence of AMF. In the presence of AMF also lower net N uptake cost was observed for the decomposition of plant-derived and microorganism-derived substrates compared to inorganic nitrogen suggesting that AMF actively controls the decomposer comunity and their carbon demand.
Conclusion
Our results identify for the first time a functional overlap of soil microorganisms as identical substrate is decomposed by different microorganisms suggesting functional redundancy of microbial communities. In consequence a better understanding of ecosystem element cycling can only be achieved when the whole plant-microorganism-organic matter-soil continuum is investigated.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Plant growth is mostly limited by nutrient availability in soil, as the major part of nutrients are not plant-available free ions but soil organic matter bound and associated with the mineral matrix (Veresoglou et al. 2012). Consequently, plants invest substantial parts of their photoassimilated carbon into soil via root exudates (sugars, amino acids and organic acids) and rhizodeposits, to stimulate microbial decomposition of organic matter in soil and nutrient release (Kuzyakov and Xu 2013; Jacoby et al. 2017). Additionally, root exudates can also directly alter organo-mineral associations (Keiluweit et al. 2015), pH (Malik et al. 2018) and redox potential (Husson 2013) of the soil to improve nutrient availability. However, immediate plant uptake of available nutrients is not always possible (Dijkstra et al. 2013), since soil microbial communities have a high nutrient demand and often assimilate nutrients more efficiently than plants (Levin and Angert 2015; Zhu et al. 2017) and plants mainly benefit in the long term from the available nutrients. Since nutrient cycles in soil are largely controlled by the various biotic (root and microbial traits) and abiotic soil properties (mineralogy, pH and redox), variations in root exudation patterns and association between plants and microorganisms can have a tremendous impact on the rates of nutrient cycling in the rhizosphere (Craine et al. 2007; Veresoglou et al. 2012).
To enhance nutrient uptake from soil a majority of terrestrial plant species (> 80%) rely on direct root association with arbuscular mycorrhizal fungi (AMF) (Bonfante and Genre 2010; Davison et al. 2015). Though AMF cannot decompose organic matter themselves (Tisserant et al. 2013), they can use their hyphal network to gain nutrients and directly compete with the decomposer community for nutrient uptake (Averill et al. 2019; Bunn et al. 2019). The AMF network also benefits plants by allowing indirect root access to a greater soil volume (Sanders and Tinker 1973), immobile nutrient pools (Hestrin et al. 2019) and distant water pockets (Bowles et al. 2018; Karlowsky et al. 2018), thus greatly increasing the area across which plant carbon is allocated. In this symbiosis plants deliver fresh photo-assimilates to AMF in exchange for nutrients. AMF grow their hypal network from the photo-assimilates and therefore have likely a high carbon demand. However the targeted allocation of plant-derived carbon to the microbial community (Whiteside et al. 2019), which stimulates decomposition and priming, may reduce the carbon cost for nutrient uptake. In contrast, in the absence of AMF-association photo-assimilated carbon is passively released to the rhizosphere and stimulates the decomposition in an untargeted way across a comparatively reduced soil volume (Kaiser et al. 2015) suggesting an overall high carbon cost. However, it remains unclear how both modes of carbon delivery into soil affect nutrient cycles and plant-microorganism nutrient exchange, and if this is affected by the decomposed substrate, i.e. plant or microbial litter and if the plant-derived carbon demand of the microbial community is changing.
Microbial decomposition of organic matter is the most fundamental process to release nutrients into soil solution (Schimel and Bennett 2004), but the activity of the soil microbial community is often limited by the availability of labile carbon (Hütsch et al. 2002). Labile carbon is dominantly provided by roots as organic root exudates like sugars, organic acids and amino acids. Root exudates can additionally directly destabilize mineral-nutrient associations and provide mineral nutrients that activate the decomposer community (Jilling et al. 2018). Root-AMF associations are also known to alter the quantity (de Graaff et al. 2010) and biochemical profile (Shi et al. 2016; Zhalnina et al. 2018) of root exudates, which feedback on the activity and the composition of the decomposer community. In addition to root exudation, nutrient content and type of organic matter that is decomposed significantly alter associated decomposer communities and their activity (Güsewell and Gessner 2009; Strickland et al. 2009; Meier et al. 2015). Recent findings based on functional analysis of rhizosphere microbial communities strongly indicate that a functionally diverse microbial community is maintained for exudate utilization in the rhizosphere compared to the bulk soil (Nuccio et al. 2020). Though exudate and litter associated shifts in the microbial community of the rhizosphere are well known, understanding the underlying variations of the carbon-nutrient exchange between plants and their fungal association and consequent variations in the decomposer communities across various nutrient sources can offer further insights into mechanisms of plant nutrient uptake and functioning of the microbial community.
Soil receives nitrogen containing substances either via carbon rich plant residues, nitrogen rich microbial residues or inorganic nitrogen from deposition and fertilizers (Gleixner 2013; Thuille et al. 2015; Liang et al. 2017). However, the difference amongst these inputs based on nutrient content (litter quality like C/N) often hides the effect of their biochemical characteristics (litter type like plant cellulose vs. microbial peptidoglycan or xanthane) on decomposer community assembly and nutrient exchange. Under natural conditions, the decomposer community is typically shaped by their biochemical and physiological capabilities, which enable them to cover their energetic and nutritional needs (Thomson et al. 2013). In consequence a substrate can be only decomposed and metabolized by organisms that have the genetic potential to produce the corresponding enzymes. Even though litter decomposition in general is well studied, the aspect of litter quality vs. litter biochemistry and follow on effects on the decomposer community structure are less understood, thus underscoring the need for further investigation through labelled nutrient amendments.
In summary, large uncertainties still exist regarding the mechanistic benefits of plant-AMF associations with respect to the role of plants and their photoassimilates in changing the microbial community functioning in organic matter decomposition and nutrient return to the plant shoot. Therefore, we assembled two plant communities that differed in their ability to either associate with AMF or not associate with AMF and amended the soil of ingrowth cores with three 15N-labeled nitrogen substrates (plant root litter, microbial necromass and inorganic N) of similar quality (C/N ratio). We traced the net plant-derived 13C uptake into the microbial community (phospholipid fatty acids (PLFA) / neutral lipid fatty acids (NLFA)) of the ingrowth core soil using a 13CO2 pulse label experiment and determined the 15N return from substrate decomposition back to the shoots. We also investigated the microbial community composition using DNA and RNA based amplicon sequencing in the ingrowth core soil in order to analyze the microbial community functioning. We first hypothesize that AMF associations result in a smaller net plant-derived carbon uptake of the microbial community (including AMF) for nitrogen mineralization in the ingrowth cores as the fungal hyphae enable target oriented exudation into the decomposition hotspots in the ingrowth cores. Second, we hypothesize that net N uptake cost is higher in presence of AMF association than absence, since AMF associations require considerable amounts of plant carbon for maintaining the hyphal network. We finally hypothesize that the associated soil microbial community in the ingrowth cores depends on the plant community (AMF vs. non-AMF) and on the substrate origin suggesting that suggesting a large functional redundancy of the microbial community.
Materials and Methods
Production of 15N labeled decomposition substrates
Plant root litter was harvested from perennial grass Holcus lanatus that was grown in a pot (57 × 37 × 20 cm) containing sand in a growth chamber. The chamber was daily lit for 12 h and had day temperatures of 23 °C and 63 % relative humidity. The plants were watered 2–3 times a week and fertilized weekly with a balanced N-free Hoagland’s solution amended with 0.40 atom% K15NO3 as the sole nitrogen source. At the end of 4 weeks the plants were harvested and roots were cleaned with distilled water. The root material was dried at 60 °C and shredded to remove effects of particle size.
Inoculum for the production of microbial necromass was generated by slow centrifugation of 6 g of Jena Experiment soil (see Microcosm Experiment section for more details) in 20 ml of distilled water at 650 g for 15 min. The supernatant was inoculated into M9 minimal medium amended with 20 % glucose and contained K15NO3 (0.40 atom%) as only nitrogen source. The culture grew for 7 days at 19 °C on a horizontal shaker. The entire cell mass was harvested by centrifugation (at 3500 g for 15 min) and was cleaned by re-suspension in 10 ml of sterile distilled water. After sterilization in an autoclave and centrifugation the resulting sterile pellet was freeze dried and homogenized for further use.
The inorganic nitrogen substrate was generated by mixing KNO3 and K15NO3 such that the mixture had an isotopic signature of 0.40 atom% and used directly for the amendments.
Microcosm Experiment
The soil used for the experiment was collected from the Jena Experiment, a grassland biodiversity experiment located in Jena, Germany in March 2015. The soil type was Eutric Fluvisol (Roscher et al. 2004) that has a pH of 7.4 (s.d. 0.06), organic carbon content of 21.1 g C kg−1 (s.d. 3.6 g C kg−1) and total nitrogen content of 2.2 g N kg−1 (s.d. 0.3 g N kg−1) (Weisser et al. 2017). The concentration of extractable nitrate and ammonium were 0.489 g N gdry soil−1 (s.d. 0.173 g N gdry soil−1) and 0.475 g N gdry soil−1 (s.d. 0.114 g N gdry soil−1) respectively. Organic phosphorous content of the soil, as measured using stepwise sequential P fractionation (Hedley et al. 1982), was 30.04 mg P kg dry soil−1 (s.d. 16.40 mg P kgdry soil−1) whereas labile inorganic phosphate content of the soil was 20.04 mg P kgdry soil−1 (s.d. 17.81 mg P kgdry soil−1) (Hacker et al. 2015). The soil contains a diverse mixture of bacteria and fungi (Weisser et al. 2017) that served as a natural inoculum in our experiment. Previous studies have demonstrated that a diverse AMF community existed at the site (Dassen et al. 2017) that proved as natural inoculum under greenhouse conditions (Dietrich et al. 2020). After removing the root mat in the upper 2 cm, the top 5 cm of soil was collected, sieved < 2 mm and all remaining roots were removed. The soil was well mixed and stored at 4 °C until the establishment of the microcosms 1 week later.
We established mixed temperate plant communities of two types namely AMF-dependent plant community (Linum usitatissimum, Linum perenne, Festuca rubra and Tagetes patula) termed as the AM plant community and plant community lacking in AMF associations (Dianthus caryophyllus, Matthiola longipetala sub-species bicornis, Dianthus barbatus and Carex arenaria) termed as NM plant community (Brundrett 2015b, 2015a). We used plant communities instead of monocultures in our experiment in order to overcome well-known species-specific effects of root exudation and consequently established 4 species mixtures, which are generally known to overcome species-specific effects (Weisser et al. 2017). In consequence, our experiment provides plant community level understanding of nutrient exchange between plants and microorganisms.
We established microcosms (19 × 12.5 × 9.5 cm) for a replicated and randomized double label experiment in the greenhouse. Each microcosm had a single central PVC ingrowth core (diameter = 5 cm), which was the primary compartment of investigation. The ingrowth cores had 8 windows (4 × 1.5 cm) guarded by a 1000 micron mesh to ensure the ingrowth of fine roots and hyphae and support the removal of the intact ingrowth core after the experiment (Figure S1). No inoculum for AMF was necessary as the source soil already contained the necessary AMF community to support the selected plant species (Harley and Harley 1987; Dietrich et al. 2020).
The microcosms and the ingrowth cores were filled with well mixed soil at field bulk density and were pre-equilibrated for 2 weeks in the greenhouse (Fig. 1). Even plant communities for each plant community type were established by planting one seedling per species. The seedlings, which had already developed first true leaves, were planted outside of the ingrowth core in each microcosm. The final plant species richness was 4 in each microcosm. The microcosms were randomly distributed in the greenhouse and maintained for 4 months until the plants were fully developed. Irrigation was daily provided for 5 min using an ebb and flood system. The plants had a daily light periode of 12 h using Son- T Agro 430 W HPS bulbs (primary light range = 520–610 nm, Philips Lighting Company, Somerset, NJ, USA). Plant species richness and evenness were visually monitored throughout the experiment.
In order to add the 15N-labeled decomposition substrates to the fully developed microcosms, the central ingrowth core was removed without disturbing the outer soil ring and root mat (Fig. 1). The soil from the ingrowth core was sieved < 2 mm removing all root material. 15N labeled substrates were added matching a total input of 550 mg C, 12 mg N and 150 µg 15N into 200 g of soil for each ingrowth core. This addition increased the carbon and nitrogen content by 13% and 2.7%, respectively. For matching the missing C we added 1,28 g and 1,58 g Xanthane, which is a N-free decomposable microbial cell wall glyco-polymer (Li et al. 2009), to the 15N labeled microbial necromass and inorganic 15N, respectively (see above). The amended soil was mixed, refilled into the ingrowth core and reinstalled on the microcosm.
Two weeks after the substrate addition, which allowed roots and hyphae to fully regrow into the ingrowth core (Figure S2), all microcosms were simultaneously exposed to a 13CO2 pulse in an air tight plexiglass chamber (Malik et al. 2015). The chamber was lit with PAR intensities above 500 µmol photons m−2 s−1 throughout the labeling. The chamber air was flushed with CO2 free synthetic air until the CO2 concentration fell below 150 ppm at the beginning of the labeling. Enriched 13CO2 (99 atom%) was introduced into the chamber using a mass flow controller (model- MF1-20sccm-N2, MKS Instruments Inc.) at a flow rate of 3.33 × 10–7 m3s−1 for 10 min and circulated using internal fans. The CO2 concentration was monitored online by a cavity ringdown spectrometer (G2101-I, Picarro Inc., Santa Clara, CA, USA) throughout the labeling period of 1 h. An additional maintenance stream of 13CO2 was introduced halfway at a flow rate of 1.67 × 10–7 m3s−1 for 5 min that kept the chamber CO2 concentration between 350 and 410 ppm. The chamber temperature was maintained close to greenhouse temperature by placing racks of frozen ice packs along the vertical walls of the chamber.
Replicated control pots (n = 3) for each substrate addition were also placed for an hour in the chambers with identical setting but using natural abundance CO2 pulsing instead of 13CO2. To avoid cross contamination from 13C labeled air, soil and biomass, the control microcosms were CO2 pulsed and harvested before the 13C labeled pots.
After 1 h of CO2 pulse the microcosms were removed from the chamber and returned back to the greenhouse. The control pots were harvested immediately for plant community analyses, microbial community composition using 16S RNA gene amplification and NLFA/PLFA extractions. The labeled plots were harvested 240 h after the 13C pulse for plant community analyses and NLFA/PLFA extractions. Previous experiments have shown that a sampling 240 h after the pulse optimizes for the 13C uptake in slow and fast parts of the microbial community using PLFA and NLFA (Malik et al. 2015; Karlowsky et al. 2018). The ingrowth core soil was removed, roots were separated by sieving to 2 mm and hand picking and the remaining rhizosphere soil was divided into three sub-samples and stored at -80 °C for molecular analysis within 1 h of harvest, dried at 105 °C for 48 h and stored at room temperature for elemental and isotope analysis and at -20 °C for microbial biomass (PLFA/NLFA). The above and below ground plant tissues were dried at 60 °C for 48 h, ball-milled to a fine powder and stored in dark at room temperature for future analysis.
Elemental and isotope analysis of plant samples
Shoot and root samples were weighed into tin capsules for total C and N content analysis using a vario EL CNS analyzer (Elementar Analysensysteme GmBH). Similarly, for bulk 13C and bulk 15 N analysis of shoot and root samples were weighed into tin capsules and analyzed using an elemental analyzer EA1100 (CE, Mainland) coupled via a conflow III interface to a DELTA + (Finnigan MAT) isotope ratio mass spectrometer (Malik et al. 2015).
Biomarker extraction, quantification and compound specific isotope analysis
To assess the net 13C uptake into the microbial community, fatty acids from neutral lipids and phospholipids were extracted from pooled ingrowth core soil, as described earlier in Karlowsky et al. 2018. In brief, 5 g of soil were extracted with a modified Bligh and Dyer (1959) lipid extraction using pressurized solvent extraction. The extract was separated using solid-phase extraction (SPE) columns (CHROMABOND SiOH, 2 g, 15 ml; Macherey–Nagel GmbH & Co. KG) into neutral lipid and phospholipid fractions. Both fractions were hydrolyzed and methylated with methanolic KOH. The resulting fatty acid methyl esters (FAMEs) were further purified using aminopropyl-modified SPE columns (CHROMABOND NH2, 0.5 g, 3 ml; Macherey–Nagel GmbH & Co. KG). The concentration and compound-specific 13C content of the samples was determined using a gas chromatograph coupled with a flame ionization detector (FID) and isotope ratio mass spectrometer (IRMS) respectively.
PLFAs were quantified on a GC-FID 7890B with a programmable temperature vaporization (PTV) injector (Agilent Technologies,.Palo Alto, CA, USA) using a DB-1MS column (60 m × 0.25 mm × 0.25 μm; Agilent Technologies) and helium as carrier gas (1.8 ml/min). The temperature program started at 45 °C for 1 min, increased at a rate of 60 °C/min to 140 °C, held for 0.5 min, increase at 2 °C/min until 264 °C and finally increased at 60°/min until 320 °C, which was held for 3 min. Directly after injection, the PTV was heated up from 55 °C to 280 °C at a rate of 500 °C/min.
NLFAs were quantified on a GC-FID HP6890 (Agilent Technologies) with constant injector temperature (280 °C), using a DB-1MS column (50 m × 0.32 mm × 0.52 μm, Agilent Technologies) and helium as carrier gas (2 ml/min). The temperature program started at 140 °C for 1 min, followed by a first ramp of 2 °C/min until 270 °C, held for 6 min and a second ramp of 30 °C/min until 340 °C, held for 5 min.
Identification of FAMEs was done by comparison of chromatograms with different known FAME mixtures (Supelco 37 Component FAME Mix; Sigma-Aldrich.Chemie GmbH; BR2 and BR4 mixture, Larodan Fine Chemicals AB) and an in house database using GC/MS (Kramer & Gleixner, 2006; Mellado-Vázquez et al., 2016; Thoms, Gattinger, Jacob, Thomas, & Gleixner, 2010).
Compound-specific 13C isotope analysis of NLFAs and PLFAs was done by GC-IRMS (GC 7890A with PTV injector; Agilent Technologies; coupled via a Conflo IV/GC IsoLink to a Delta V Plus IRMS; Thermo Fisher Scientific) using a DB-1MS column (60 m × 0.25 mm × 0.25 μm; Agilent Technologies) and helium as carrier gas (1.8 ml/min). Directly after injection, the PTV was heated up from 55 °C to 280 °C at a rate of 500 °C/min. The GC temperature program started at 45 °C for 1 min, then increased in a first ramp of 60 °C/min to 140 °C (held for.0.5 min), followed by a second ramp of 4 °C/min until 283 °C (held for 4.9 min) and a third ramp until 320 °C (held for 3 min). Concentrations and 13C isotope content of identified FAMEs were corrected for the methyl group introduced during derivatization.We summed the FAMEs 14:0, 15:0, 16:0, 17:0, 18:0, 14:0i, 15:0i, 15:0a, 16:0i, 17:0i, 17:0a, 16:0(10Me), 17:0(10Me), 18:0(10Me), 17:0cy, 19:0cy, 15:1, 16:1ω7, 16:1ω5, 16:1, 17:1, 18:1ω7 and 18:1ω9 from the PLFA fraction as bacterial markers, FAME 18:2ω6,9 from the PLFA fraction as marker for free-living fungi and FAME 16:1ω5 from the NLFA fraction as marker for AMF (Karlowsky et al. 2018). The yield (volume x concentration) of saprotrophic fungal markers (PLFA 18:2ω6,9) were converted into biomass C by applying the conversion factor 11.8 nmol = 1 mg C (Klamer 2004), for bacterial biomarkers by applying conversion factor 363.3 nmol = 1 mg C (Frostegård and Bååth 1996) and for 16:1ω5 NLFA by applying conversion factor 1.047 nmol = 1 μg C (Williams et al. 2017). All values are corrected for soil moisture and the total weight of ingrowth core soil.
The net mass of isotopes (13C (µg) and 15N (µg)) assimilated into biomass and its fractions (shoot, total microbial biomass bacteria, fungi and AMF) (µg) was determined from the difference between the isotope content in the samples (atom%labelled) collected at the end of the experiment and the control samples (atom%control) times the total pool size (Cpool, Npool), which integrates over the total soil mass of the ingrowth core and community biomass for plants and microbial fractions (Karlowsky et al. 2018).
The net N uptake cost or more specific the net plant C allocation to microbial biomass and groups per nitrogen gain in the shoot (µg/µg) was calculated dividing the net 13C assimilated (incorporated 13C (µg)) of the soil microbial community and its parts (bacteria, fungi and AMF) by the incorporated 15N (µg) into the shoots.
DNA and RNA based microbial community profiling at the time of the labeling
To determine the microbial community composition, 1.09 ± 0.01 g of soil from each treatment was used to co-extract total RNA and DNA using the MoBio RNA Powersoil kit. The samples were bead-beaten for one cycle at 5.5 m/s for 30 s with a FastPrep instrument (MP Biomedicals Germany GmbH). DNA was extracted using a MoBio RNA Powersoil DNA elution accessory kit. Traces of DNA that remained in the final RNA sample were removed using a MoBio DNase Max kit. The concentrations of purified DNA and RNA were measured using Qubit 4 fluorometer (Thermo Fisher Scientific) using the broad range assay kits for DNA and RNA. Additionally, the purity of the samples was checked using a nanodrop spectrometer 2000c (Thermo Fisher Scientific). All samples had A260/280 and A260/230 ratios of 2 suggesting that the samples had no protein or organic matter contamination. The integrity of the RNA samples was visually verified using 0.8% agarose gel electrophoresis. Presence of sharp well-defined ribosomal RNA bands and smear in between and above these bands was considered as an indicator of high integrity of the RNA. One replicate of the AM plant community amended with plant root litter and microbial necromass was discarded due to poor RNA integrity and subsequent non-amplification of cDNA. 200 ng of total RNA extracted was converted into cDNA using Superscript™ VILO kit (Thermo Fisher Scientific) using random hexamers to ensure uniform conversion of all the RNA fragments. The DNA and cDNA samples were further used for amplifying the 16S rRNA gene and ITS regions respectively. Briefly, for the 16S rRNA gene sequencing, amplicon libraries were prepared with V3-V5 primers (Haas et al. 2011) using PCR conditions described by Kozich et al. 2013. Three samples without amplification of extracted DNA were discarded from further investigation (AM soil amended with plant litter and microbial necromass and NM soil amended with inorganic N). ITS2 gene libraries were constructed by amplifying ITS region 2 using the fITS7 (forward) and ITS4 (reverse) primers (Ihrmark et al. 2012) with previously described PCR conditions (Gweon et al. 2015). Both sets of primers also consisted of an appropriate Illumina adapter, an 8‐nt index sequence, a 10‐nt pad sequence, a 2‐nt linker (Kozich et al. 2013). The amplicons were sequenced using Illumina MiSeq, v3 chemistry, 600 cycles.
Processing of sequencing data
For the bacterial community 16S rRNA gene sequencing data, the forward and reverse reads obtained were merged using PEAR (v 0.9.6) with default parameters (Zhang et al. 2014). The bacterial community composition was analyzed using the QIIME (1.8.0) pipeline (Caporaso et al. 2010). Representative OTU’s were picked using UCLUST using de novo method at 97% similarity level and were assigned taxonomy using RDP classifier method with the SILVA reference data set (release 132 for QIIME) (Wang et al. 2007; Edgar 2010). Total number of sequencing reads classified within the OTU table for the samples varied between 4000 to 21,982 reads. In order to make inter-sample comparisons the OTU table obtained was rarefied to 4000 sequences per sample using QIIME command single_rarefraction.py (Caporaso et al. 2010).
Since fungal spores are ubiquitous, the ITS2 regions that were transcribed into RNA were used to sample transcriptionally active fungi (Anderson and Parkin 2007). The amplified ITS region from total cDNA samples were used for sequencing. The amplified ITS sequencing data was analyzed using the PIPITS pipeline using the default parameters (Gweon et al. 2015). Briefly, the paired end sequences were merged using VSEARCH and the ITS2 region was extracted using ITSx. The ITS2 region was matched to a trained UNITE database at 97% similarity using RDP classifier to obtain an OTU table (Bengtsson-Palme et al. 2013; Rognes et al. 2016). The OTUs were assigned to fungal families using FUNguild assignment app (http://www.stbates.org/guilds/app.php) (Nguyen et al. 2016).
The bacterial and fungal amplicon sequencing data and the associated metadata is publicly available through the Sequence Read Archive project number: SRP132171. A detailed table of associated metadata and download links is also provided in the supplementary information (Table S12 and S13).
Statistical data analysis
One-way ANOVA was used to determine statistical differences amongst rhizosphere types (AM vs. NM) for root to shoot ratio, 15N assimilated by plant shoot and net microbial 13C assimilation into total microbial biomass. Two-way ANOVA was applied to identify statistically significant differences for plant carbon allocation per unit 15N gain due to rhizosphere and litter types. All two-way ANOVA models that showed significant interactions of rhizosphere-litter types were subject to Tukey’s post-hoc test to identify differences amongst litter types nested within rhizosphere types. Significant differences were only considered if the assumption for homogeneity of variance was met using the Flinger-Killing test (Table S2). Subsampled OTU tables were binned to different taxonomic groups at the phylum and class levels (bacteria) whereas fungal OTUs were binned into guilds to compare effect of AM vs. NM plant communities and substrate amendments on microbial (bacterial and fungal) community composition. The effects of substrate addition and plant community on the biomass of specific microbial groups were determined using a two-way ANOVA on their relative abundance. Changes in the microbial (bacterial and fungal) community composition by the plant community and the substrates addition were analyzed using non-parametric Permutational Multivariate Analysis of Variance PERMANOVA (“adonis” function in the vegan package 2.5–2) (Oksanen et al. 2018). The assumption of homogeneity of variance was tested using the betadisper function (vegan package 2.5–2) and both the fungal and bacterial community composition was found to meet this assumption (Table S9). We visualized the dissimilarities between the microbial communities under the influence of plant communities and substrate added to the soil using non-metric multidimensional scaling (NMDS) of Bray–Curtis dissimilarity coefficients. All analysis was performed using R software (v 3.4.3) with visualization being done with ggplot2 package (Wickham 2011).
Results
Carbon allocation and Nitrogen return
The AM plant community had expectedly a lower shoot biomass (4.62 ± 0.14 g) compared to NM the plant community (7.42 ± 0.44 g) but both had similar root biomass (F1,16 = 0.17; p = 0.685), which resulted in a higher root to shoot ratio for the AM plant community (data not shown). In line with this the AM plant community also allocated more carbon to their roots, which was reflected in the root to shoot ratio of incorporated 13C in the AM plant community compared to the NM plant community (F1,16 = 20.91, p < 0.001; Fig. 2A). In contrast, the net 13C assimilation of the rhizosphere microbial communities, which decomposed the added substrates in the ingrowth cores of the AM plant community, was significantly lower than that of the NM plant community (F1,16 = 21.75, p < 0.001, Fig. 2B). This was supported by lower 13C assimilation into total microbial biomass (CFE) and respired soil CO2 (data not shown).
Photosynthetically fixed carbon assimilated by plants and associated microbial communities. A: Allocation of 13C between roots and shoots of AM (n = 9) and NM (n = 9) plant communities across all soil amendments. B: Mean 13C excess (µg 13C g−1 soilDW) in the soil microbial biomass of each plant community types. Height of the bars represents mean of all substrate additions and error bars represent standard error of mean. Different letters on bars designate significant differences based on one-way analysis of variance (ANOVA) comparing both plant communities
The shoots of the AM plant community gained 93.1 ± 4.7 μg 15N from labeled substrate decomposition, which was not significantly different from the NM plant community that gained 82.3 ± 10.7 μg 15 N ((F1,16 = 0.40, p = 0.541, Fig. 3A). The 15N uptake of the plant community shoots was also not affected by the added substrate (AM: F2,6 = 0.57, p = 0.594; NM: F2,6 = 0.797, p = 0.493). Unexpectedly, the net N uptake cost in the AM plant community was significantly lower than in the NM plant community (F1,16 = 15.20, p = 0.001; Fig. 3B), as 66% less 13C was net assimilated by the microbial community per 15N gain of the AMF plant community shoots. Even though, the AM plant community allocated significantly more 13C into AMF biomass per 15N gain of the plant shoot (F1,16 = 30.53, p < 0.001; Fig. 4A) this was overcompensated by a significantly lower 13C allocation into free-living fungi (F1,16 = 15.47, p = 0.001; Fig. 4B) and bacteria (F1,16 = 4.78, p = 0.044; Fig. 4C) per 15N gain of the plant shoot. Interestingly, the net N uptake cost depended only in the AM plant community also on the decomposed substrate (F2,15 = 7.13, p = 0.009, Fig. 4). Specifically, in the AM community resulted the colonization of ingrowth cores with inorganic N amended soil in significantly higher net 13C allocation to bacteria and free-living fungi compared to microbial necromass and plant litter containing ingrowth cores (Fig. 4B, C). Such substrate-based differences were missing in the ingrowth cores of NM communities.
Carbon investment into microbial community and return of decomposition released nitrogen. A: 15N content (µg) of shoots in AM (n = 9) and NM (n = 9) plant communities. B: Net N uptake cost (µg 13C in microbial biomass / µg 15N plant shoot) Colors represent plant community types (Dark Green = AM and Orange = NM plant community). Horizontal bars represent means for all substrate additions and error bars represent standard error of means. Groups sharing the same letters are not significantly different based on one-way analysis of variance (ANOVA)
Net N uptake cost for different microbial groups. Mean 13C (µg) incorporated into total microbial group specific lipids (A – AMF (NLFA); B – Free-living fungi (PLFA) and C – Bacteria (PLFA)) per 15N uptake (µg) by the plant community shoots. Means, standard error and significant differences between plant communities (asterisk) and soil amendments (LSD letter-group based on Tukey’s post-hoc tests) are shown. Significance is indicated at p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***).
Microbial community composition
The microbial community that developed in the ingrowth cores of AM and NM plant communities, had the same OTU richness and evenness (Table S3). Only the addition of root litter significantly increased the OTU richness of fungi in the ingrowth cores (Table S4). In contrast, the relative abundance of individual bacterial groups and fungal guilds significantly differed between the ingrowth cores of the AM and NM plant community and with the added substrate (plant root litter, microbial necromass and inorganic-N) (Fig. 5, Table S10 and S11). Fungal and bacterial communities are mainly separated by the added substrate on NMDS1 whereas AM and NM plant community separated on NMDS2. The microbial communities after root litter addition were more dissimilar to the other substrate addition, which is likely caused by the presence of plant carbohydrates as C substrate vs. microbial carbohydrates that were added for microbial necromass and inorganic N additions.
Variation in microbial bacterial and fungal community composition with different plant communities and added substrates. Non-metric multidimensional scaling (NMDS) plots illustrating Bray–Curtis distances between microbial communities (A: Fungal Community (ITS regions) and B: Bacterial Community (16S RNA gene) in individual samples (n = 18). Colors represent plant community type (Dark Green = AM and Orange = NM) whereas symbols represents soil amendments. Stress values for fungal (A) and bacterial (B) plots are 0.05 and 0.18 respectively. Fungal and bacterial communities differ significantly between plant communities (F = 1.87, p = 0.013 and F = 1.24, p = 0.019) and added substrates (F = 1.99, p = 0.0003 and F = 1.5, p = 0.0001), respectively (see Text and Table S10 and S11).
AMF associations had compared to NM communities significantly higher relative abundance of bacterial groups belonging to Actinobacteria and Proteobacteria (specifically Betaproteobacteria) (Table S5) and the absence of AMF associations had significantly higher relative abundance of the Acidobacteria group (Table S5). In agreement with results from PLFA analysis, AMF association had higher relative abundance of AMF (Table S6) predominantly belonging to Diversiporaceae, Claroidegloeraceae, Glomeraceae and Paraglomeraceae whereas NM plant communities had significantly higher relative abundance of fungal saprotrophs and endophytes that are also reported partially having a free-living lifestyle (Rodriguez et al. 2009).
The response of the bacterial community on substrate additions was generally higher compared to the fungal community (Fig. 5, Table S7 and S8). The substrate addition of root litter had a stimulating effect on the relative abundance bacterial groups belonging to Gemmamonadetes, Nitrospira and others whereas the addition of bacterial necromass showed significantly lower abundance of Deltaproteobacteria (Table S7). Only fungal OTUs that were not exclusively assigned to major guilds (Saprotrophs, AMF, Ectomycorrhizal and Endophytes) showed a significant response to substrate amendments (Table S8).
Discussion
In this study, we investigated the role of plants in ecosystem nutrient cycling by tracing plant-derived carbon in substrate decomposing microorganisms and substrate-derived nitrogen back to plant shoots. We compared on the one hand a plant community with and one without symbiotic fungi (AMF) and on the other hand the decomposition of different substrates. We investigated the changes in the microbial community composition and determined the net plant-derived carbon assimilation of the microbial communities and their functional parts (bacterial groups and fungi). Finally we calculated the net carbon cost for decomposition-derived nitrogen uptake to the plant shoots. We found that AMF associated (AM) plant communities did not gain significantly more nitrogen from the different substrates than non-mycorrhizal (NM) plant communities. This is in line with previous reports looking at nitrogen uptake on the plant community level (van der Heijden et al. 2006). Even so, it is clear from experiments on the species level using compartments with restricted hyphal access (Nuccio et al. 2013) and sterile soil (van der Heijden et al. 2006) that plants clearly benefits from AMF for nutrient uptake. Our results suggest therefore that results from single species experiments cannot be generalized for plant communities as associations in single species experiments often occur with specialized AMF species and do not account for alternate nutrient uptake strategies that exist in NM plants. We advocate consequently community-based approaches to understand effects of presence or absence of AMF association. We used four different plant species in our experimental plant communities, as biodiversity research has shown that species related trait variations are less important in plant communities having 4 and more species (Weisser et al. 2017).
Most interestingly, our experiments show that even though AMF associated plants may take up nutrients more efficiently, the total nutrient gain of the plant community relative to NM based plant communities remains similar. This raises the question why so many terrestrial plant species rely on the AMF association? At first glance, it appears that AMF associated plants need to allocate carbon to grow and maintain the hyphal network (Kaiser et al. 2015), which is theoretically increasing the plant carbon demand. In addition AM plants directly exchange carbon for nutrients with the AMF. However, carbon demands are also observed in NM plants, as NM plants release exudates to induce growth of saprotrophic fungi and bacteria in the rhizosphere, which stimulate substrate decomposition. Our results (Fig. 3B, Fig. 6,) show that the targeted transfer of plant carbon to AMF and from AMF directly to the location of microbial decomposition is more efficient than untargeted transfer of carbon to the whole rhizosphere of NM roots. This target oriented carbon transfer likely explains the overall success of plant-AMF associations. This is also in line with previous studies showing the importance of carbon flow through the AMF hyphal network for functioning of the associated soil bacterial community (Vestergård et al. 2008; Drigo et al. 2010; Kaiser et al. 2015). The close interaction of AMF and bacteria potentially increases decomposition efficiency and makes in the long run more nutrients available to the plant communities (Verbruggen et al. 2013). NM plant communities likely miss the targeted growth of AMF hyphae and rely on free-living saprophytic fungi and bacteria for decomposition, therefore increasing the overall carbon per nitrogen cost (Cosme et al. 2018).
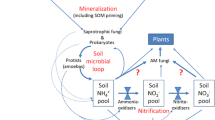
Conceptual model showing the effect of AMF association on decomposer and plant responsive microbial groups that shape carbon and nutrient assimilation when decomposing stoichiometrically identical substrates. In non-mycorrhizal plants (NM) the role of AMF is taken over by fungal saprotrophs. Distinct bacterial communities are assembled for each substrate depending on substrate origin and plant-AMF association context. Arrows represent the C and N flow differences arising from presence or absence of AMF, explaining that for an equivalent N uptake AMF associated plants invested lower amounts of photoassimilated carbon into decomposer community
Belowground plant carbon allocation into the microbial community is one of the most important mechanism for soil carbon formation and nutrient cycling (Gleixner 2013; Lange et al. 2015; Walker et al. 2020). Plants adjust the below ground carbon flow and its chemical composition in response to the environment. For instance, increased fluxes of rhizodeposits that stimulate decomposition are observed as a response to increased atmospheric carbon concentrations in order to compensate to the higher nutrient demand for growth (Drigo et al. 2010). Also the the nutrient content (C/N ratio) of organic matter is suggested to influence the flux and composition of rhizodeposites (Jones et al. 2004; Wang et al. 2015; Stempfhuber et al. 2016). In our experiment we added the same amount of carbon and nitrogen for microbial decomposition, but varied the biochemistry of the substrates. Root litter held nitrogen mainly as proteins, microbial necromass mainly as peptidoglucanes and nitrate as third substrate was readily available. Our substrate addition showed that the net assimilation of plant-derived carbon by the rhizosphere microbial community depended on the biochemical characteristics of the decomposed substrate (Fig. 4). This strongly suggests that substrate (bio)chemistry and therefore the enzymes that are needed for the decomposition of the substrates and the microorganisms that are able to biosynthesize them, is likely the reason behind the effect of nutrient content (C/N ration) in substrate decomposition experiments. Therefore, our results advocate that future experiments that investigate the below ground carbon transfer and decomposition should simultaneously consider nutrient content (C/N ratio) and biochemistry of substrates available for nutrient recycling.
AMF associated rhizosphere microbial communities generally assimilated less plant carbon than the microbial communities lacking this association (Fig. 4), which suggests that AMF allocated less labile carbon to the free-living soil microbial community in order to take up mineralized N from decomposition. The fundamental role of the AMF in this transfer is supported by results from a root exclusion treatment that used 30 µm mesh instead of 1000 µm (Table S1), which showed no significantly different 15N uptake into the plant community. This suggests that AMF indeed act as target-oriented root extensions that more effectively feed plant-derived carbon to the free-living microbial community (Parihar et al. 2020) and therefore likely use the flux and composition of exudates to stimulate the suitable part of the microbial community for decomposition of the available substrate. This targeted feeding strategy is supported by the significant response of specific microbial groups to either the substrate addition or the plant community (AM and NM) (Fig. 5).
Therefore, the composition of the microbial decomposer community is adjusted to plant exudation and type of the substrate being decomposed (Fig. 5, Fig. 6), thus making AMF associations with plants an important part of the decomposition process (McLaren and Turkington 2011). Expectedly, the fungal community primarily differed between AM or NM plant communities (Fig. 5A), emphasizing the fungal role in decomposition and nutrient gathering (Meier et al. 2015; Bukovská et al. 2018). However, in the presence of AMF, we observed lower plant carbon assimilation by the free-living fungal community (Fig. 4B) (Mueller et al. 2014), indicating dominance of a bacterial community in the associated decomposition processes. This suggests that primary decomposers in the AM rhizosphere/hyphosphere were bacteria that were fed and rapidly adapted to substrate by the AMF (Herman et al. 2012; Nuccio et al. 2013).
Most interestingly our results indicate for the first time that AM plant communities strongly benefit from the association with AMF during nutrient uptake from microbial necromass (Fig. 4). AMF assimilated more 13C from plants (i.e. higher net N uptake cost, Fig. 4A) but invested less carbon into the bacterial community for decomposition (i.e. lower net N uptake cost, Fig. 4B). This might indicate that AMF, which lack capability to decompose lignocellulose structures of plant cell walls themselves (Tisserant et al. 2013), might be able to decompose peptidoglycan structures of microbial cell walls. This idea would be supported by the dramatic up-regulation of genes in AMF that encode for enzymes containing a peptidoglycan binding LysM domain and glycan hydrolases like lysozyme, glucosamidase and alpha amylase, which would be necessary for microbial cell wall decomposition (Tisserant et al. 2013; Gude et al. 2012). If we consider that large parts of soil organic matter are microbial necromass (Gleixner 2013) the association of plants with AMF gain a new overall importance for ecosystem functioning. However, additional experiments are needed to confirm this new role of plant-AMF associations for microbial cell wall decomposition.
Conclusion
Our results suggest that a three-way interaction between the plant community, the microbial community and the biochemistry of decomposed substrate is of fundamental importance for ecosystem functioning. Specifically, nutrient recycling in the plant-microorganism-soil continuum is shaped by these interactions leading to a plant-driven and context-dependent (biochemistry of litter, environmental conditions, presence or absence of fungi) stimulation of microorganisms that decompose the given substrate (Fig. 6). Most interestingly is the decomposition capability of parts of the decomposer community likely redundant as litter with the same biochemistry is decomposed by different parts of the microbial community in dependence of the presence and absence of AMF (functional overlap). Consequently, organic matter decomposition and nutrient recycling can only be fully understood if the whole continuum is investigated. Moreover, our results demonstrate for the first time that plant nutrient gain via AMF association is less carbon intensive and likely more targeted than passive exudation of plant communities lacking an AMF association. This might explain why a large part of the plant community relies on AMF mediated nutrient uptake strategy.
References
Anderson IC, Parkin PI (2007) Detection of active soil fungi by RT-PCR amplification of precursor rRNA molecules. J Microbiol Methods 68:248–253. https://doi.org/10.1016/j.mimet.2006.08.005
Averill C, Bhatnagar JM, Dietze MC, Pearse WD, Kivlin SN (2019) Global imprint of mycorrhizal fungi on whole-plant nutrient economics. Proc Natl Acad Sci U S A 116:23163–23168. https://doi.org/10.1073/pnas.1906655116
Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A et al (2013) Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4:914–919. https://doi.org/10.1111/2041-210X.12073
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. https://doi.org/10.1139/y59-099
Bonfante P, Genre A (2010) Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat Commun 1:48. https://doi.org/10.1038/ncomms1046
Bowles TM, Jackson LE, Cavagnaro TR (2018) Mycorrhizal fungi enhance plant nutrient acquisition and modulate nitrogen loss with variable water regimes. Glob Chang Biol 24:e171–e182. https://doi.org/10.1111/gcb.13884
Brundrett M (2015a). Arbuscular Mycorrhizas. In: Mycorrhizal Associations: The Web Resource. Version 2.0. 24.04.2015a.‹mycorrhizas.info›. Available at: http://mycorrhizas.info/vam.html.
Brundrett M (2015b). Mycorrhizal Associations: The Web Resource. Version 2.0. 24.04.2015b. ‹mycorrhizas.info›. Available at: http://mycorrhizas.info/nmplants.html
Bukovská P, Bonkowski M, Konvalinková T, Beskid O, Hujslová M, Püschel D et al (2018) Utilization of organic nitrogen by arbuscular mycorrhizal fungi—is there a specific role for protists and ammonia oxidizers? Mycorrhiza 28:269–283. https://doi.org/10.1007/s00572-018-0825-0
Bunn RA, Simpson DT, Bullington LS, Lekberg Y, Janos DP (2019). Revisiting the ‘direct mineral cycling’ hypothesis: arbuscular mycorrhizal fungi colonize leaf litter, but why? ISME J 1. https://doi.org/10.1038/s41396-019-0403-2
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Cosme M, Fernández I, Van der Heijden MGA, Pieterse CMJ (2018) Non-Mycorrhizal Plants: The Exceptions that Prove the Rule. Trends Plant Sci 23:577–587. https://doi.org/10.1016/j.tplants.2018.04.004
Craine JM, Morrow C, Fierer N (2007) Microbial nitrogen limitation increases decomposition. Ecology 88:2105–2113. https://doi.org/10.1890/06-1847.1
Dassen S, Cortois R, Martens H, de Hollander M, Kowalchuk GA, van der Putten WH et al (2017) Differential responses of soil bacteria, fungi, archaea and protists to plant species richness and plant functional group identity. Mol Ecol 26:4085–4098. https://doi.org/10.1111/mec.14175
Davison J, Moora M, Opik M, Adholeya A, Ainsaar L, Ba A et al (2015) Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349:970–973. https://doi.org/10.1126/science.aab1161
de Graaff M-A, Classen AT, Castro HF, Schadt CW (2010) Labile soil carbon inputs mediate the soil microbial community composition and plant residue decomposition rates. New Phytol 188:1055–1064. https://doi.org/10.1111/j.1469-8137.2010.03427.x
Dietrich P, Roscher C, Clark AT, Eisenhauer N, Schmid B, Wagg C (2020) Diverse plant mixtures sustain a greater arbuscular mycorrhizal fungi spore viability than monocultures after 12 years. J Plant Ecol 13:478–488. https://doi.org/10.1093/jpe/rtaa037
Dijkstra FA, Carrillo Y, Pendall E, Morgan JA (2013) Rhizosphere priming: a nutrient perspective. Front Microbiol 4:216. https://doi.org/10.3389/fmicb.2013.00216
Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ et al (2010) Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc Natl Acad Sci U S A 107:10938–10942. https://doi.org/10.1073/pnas.0912421107
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Frostegård A, Bååth E (1996) The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils 22:59–65. https://doi.org/10.1007/s003740050076
Gleixner G (2013) Soil organic matter dynamics: a biological perspective derived from the use of compound-specific isotopes studies. Ecol Res 28:683–695. https://doi.org/10.1007/s11284-012-1022-9
Gude A, Kandeler E, Gleixner G (2012) Input related microbial carbon dynamic of soil organic matter in particle size fractions. Soil Biology & Biochemistry 47:209–219. https://doi.org/10.1016/j.soilbio.2012.01.003
Güsewell S, Gessner MO (2009) N: P ratios influence litter decomposition and colonization by fungi and bacteria in microcosms. Funct Ecol 23:211–219. https://doi.org/10.1111/j.1365-2435.2008.01478.x
Gweon HS, Oliver A, Taylor J, Booth T, Gibbs M, Read DS et al (2015) PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol Evol 6:973–980. https://doi.org/10.1111/2041-210X.12399
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. https://doi.org/10.1101/gr.112730.110
Hacker N, Ebeling A, Gessler A, Gleixner G, González Macé O, de Kroon H et al (2015) Plant diversity shapes microbe-rhizosphere effects on P mobilisation from organic matter in soil. Ecol Lett 18:1356–1365. https://doi.org/10.1111/ele.12530
Harley JL, Harley EL (1987) A Checklist of Mycorrhiza in the British Flora. New Phytol 105:1–102. https://doi.org/10.1111/j.1469-8137.1987.tb00674.x
Hedley MJ, Stewart JWB, Chauhan BS (1982) Changes in Inorganic and Organic Soil Phosphorus Fractions Induced by Cultivation Practices and by Laboratory Incubations. Soil Sci Soc Am J 46:970–976. https://doi.org/10.2136/sssaj1982.03615995004600050017x
Herman DJ, Firestone MK, Nuccio E, Hodge A (2012) Interactions between an arbuscular mycorrhizal fungus and a soil microbial community mediating litter decomposition. FEMS Microbiol Ecol 80:236–247. https://doi.org/10.1111/j.1574-6941.2011.01292.x
Hestrin R, Hammer EC, Mueller CW, Lehmann J (2019) Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Commun Biol 2:233. https://doi.org/10.1038/s42003-019-0481-8
Husson O (2013) Redox potential (Eh) and pH as drivers of soil/plant/microorganism systems: A transdisciplinary overview pointing to integrative opportunities for agronomy. Plant Soil 362:389–417. https://doi.org/10.1007/s11104-012-1429-7
Hütsch BW, Augustin J, Merbach W (2002) Plant rhizodeposition — an important source for carbon turnover in soils. J Plant Nutr Soil Sci 165:397. https://doi.org/10.1002/1522-2624(200208)165:4%3c397::AID-JPLN397%3e3.0.CO;2-C
Ihrmark K, Bödeker ITM, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J et al (2012) New primers to amplify the fungal ITS2 region - evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol 82:666–677. https://doi.org/10.1111/j.1574-6941.2012.01437.x
Jacoby R, Peukert M, Succurro A, Koprivova A, Kopriva S (2017) The Role of Soil Microorganisms in Plant Mineral Nutrition-Current Knowledge and Future Directions. Front Plant Sci 8:1617. https://doi.org/10.3389/fpls.2017.01617
Jilling A, Keiluweit M, Contosta AR, Frey S, Schimel J, Schnecker J et al (2018) Minerals in the rhizosphere: overlooked mediators of soil nitrogen availability to plants and microbes. Biogeochemistry 139:103–122. https://doi.org/10.1007/s10533-018-0459-5
Jones DL, Hodge A, Kuzyakov Y (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163:459–480. https://doi.org/10.1111/j.1469-8137.2004.01130.x
Kaiser C, Kilburn MR, Clode PL, Fuchslueger L, Koranda M, Cliff JB et al. (2015). Exploring the transfer of recent plant photosynthates to soil microbes: Mycorrhizal pathway vs direct root exudation. New Phytol 205. https://doi.org/10.1111/nph.13138
Kallenbach CM, Frey SD, Grandy AS (2016) Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat Commun 7:13630. https://doi.org/10.1038/ncomms13630
Karlowsky S, Augusti A, Ingrisch J, Hasibeder R, Lange M, Lavorel S et al (2018) Land use in mountain grasslands alters drought response and recovery of carbon allocation and plant-microbial interactions. J Ecol 106:1230–1243. https://doi.org/10.1111/1365-2745.12910
Keiluweit M, Bougoure JJ, Nico PS, Pett-Ridge J, Weber PK, Kleber M (2015) Mineral protection of soil carbon counteracted by root exudates. Nat Clim Chang 5:588–595. https://doi.org/10.1038/nclimate2580
Klamer M (2004) Estimation of conversion factors for fungal biomass determination in compost using ergosterol and PLFA 18:2ω6,9. Soil Biol Biochem 36:57–65. https://doi.org/10.1016/j.soilbio.2003.08.019
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Kuzyakov Y, Xu X (2013) Competition between roots and microorganisms for nitrogen: mechanisms and ecological relevance. New Phytol 198:656–669. https://doi.org/10.1111/nph.12235
Lange M, Eisenhauer N, Sierra CA, Bessler H, Engels C, Griffiths RI et al (2015) Plant diversity increases soil microbial activity and soil carbon storage. Nat Commun 6:6707. https://doi.org/10.1038/ncomms7707
Levin PA, Angert ER (2015) Small but mighty: Cell size and bacteria. Cold Spring Harb Perspect Biol 7:1–11. https://doi.org/10.1101/cshperspect.a019216
Li B, Guo J, Chen W et al. (2009) Endoxanthanase, a Novel β-d-Glucanase Hydrolyzing Backbone Linkage of Intact Xanthan from Newly Isolated Microbacterium sp. XT11. Appl Biochem Biotechnol 159, 24–32. 10.1007/s12010-008-8439-1
Liang C, Schimel JP, Jastrow JD (2017) The importance of anabolism in microbial control over soil carbon storage. Nat Microbiol 2:17105. https://doi.org/10.1038/nmicrobiol.2017.105
Malik AA, Dannert H, Griffiths RI, Thomson BC, Gleixner G (2015) Rhizosphere bacterial carbon turnover is higher in nucleic acids than membrane lipids: Implications for understanding soil carbon cycling. Front Microbiol 6:268. https://doi.org/10.3389/fmicb.2015.00268
Malik AA, Puissant J, Buckeridge KM, Goodall T, Jehmlich N, Chowdhury S et al (2018) Land use driven change in soil pH affects microbial carbon cycling processes. Nat Commun 9:1–10. https://doi.org/10.1038/s41467-018-05980-1
McLaren JR, Turkington R (2011) Plant identity influences decomposition through more than one mechanism. PLoS ONE 6:e23702. https://doi.org/10.1371/journal.pone.0023702
Meier IC, Pritchard SG, Brzostek ER, McCormack ML, Phillips RP (2015). The rhizosphere and hyphosphere differ in their impacts on carbon and nitrogen cycling in forests exposed to elevated CO2. New Phytol 205, 1164–74. https://doi.org/10.1111/nph.13122.
Mueller RC, Balasch MM, Kuske CR (2014) Contrasting soil fungal community responses to experimental nitrogen addition using the large subunit rRNA taxonomic marker and cellobiohydrolase I functional marker. Mol Ecol 23:4406–4417. https://doi.org/10.1111/mec.12858
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J et al (2016) FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248. https://doi.org/10.1016/J.FUNECO.2015.06.006
Nuccio EE, Hodge A, Pett-Ridge J, Herman DJ, Weber PK, Firestone MK (2013) An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ Microbiol 15:1870–1881. https://doi.org/10.1111/1462-2920.12081
Nuccio EE, Starr E, Karaoz U, Brodie EL, Zhou J, Tringe SG et al (2020) Niche differentiation is spatially and temporally regulated in the rhizosphere. ISME J 14:999–1014. https://doi.org/10.1038/s41396-019-0582-x
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D et al. (2018). vegan: Community Ecology Package. Available at: https://cran.r-project.org/package=vegan.
Parihar M, Rakshit A, Meena VS, Gupta VK, Rana K, Choudhary M et al (2020) The potential of arbuscular mycorrhizal fungi in C cycling: a review. Arch Microbiol 202:1581–1596. https://doi.org/10.1007/s00203-020-01915-x
Rodriguez RJ, White JF Jr, Arnold AE, Redman RS (2009) Fungal endophytes: diversity and functional roles. New Phytol 182:314–330. https://doi.org/10.1111/j.1469-8137.2009.02773.x
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Roscher C, Schumacher J, Baade J, Wilcke W, Gleixner G, Weisser WW et al (2004) The role of biodiversity for element cycling and trophic interactions: an experimental approach in a grassland community. Basic Appl Ecol 5:107–121. https://doi.org/10.1078/1439-1791-00216
Sanders FE, Tinker PB (1973) Phosphate flow into mycorrhizal roots. Pestic Sci 4:385–395. https://doi.org/10.1002/ps.2780040316
Schimel JP, Bennett J (2004) Nitrogen mineralization: Challenges of a changing paradigm. Ecology 85:591–602. https://doi.org/10.1890/03-8002
Shi S, Nuccio EE, Shi ZJ, He Z, Zhou J, Firestone MK (2016) The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol Lett 19:926–936. https://doi.org/10.1111/ele.12630
Stempfhuber B, Richter-Heitmann T, Regan KM, Kölbl A, Wüst PK, Marhan S et al (2016) Spatial interaction of archaeal ammonia-oxidizers and nitrite-oxidizing bacteria in an unfertilized grassland soil. Front Microbiol 6:1567. https://doi.org/10.3389/fmicb.2015.01567
Strickland MS, Osburn E, Lauber C, Fierer N, Bradford MA (2009) Litter quality is in the eye of the beholder: Initial decomposition rates as a function of inoculum characteristics. Funct Ecol 23:627–636. https://doi.org/10.1111/j.1365-2435.2008.01515.x
Thomson BC, Ostle NJ, McNamara NP, Oakley S, Whiteley AS, Bailey MJ et al (2013) Plant soil interactions alter carbon cycling in an upland grassland soil. Front Microbiol 4:253. https://doi.org/10.3389/fmicb.2013.00253
Thuille A, Laufer J, Höhl C, Gleixner G (2015) Carbon quality affects the nitrogen partitioning between plants and soil microorganisms. Soil Biol Biochem 81:266–274. https://doi.org/10.1016/j.soilbio.2014.11.024
Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R et al (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci 110:20117–20122. https://doi.org/10.1073/pnas.1313452110
van der Heijden MGA, Streitwolf-Engel R, Riedl R, Siegrist S, Neudecker A, Ineichen K et al (2006) The mycorrhizal contribution to plant productivity, plant nutrition and soil structure in experimental grassland. New Phytol 172:739–752. https://doi.org/10.1111/j.1469-8137.2006.01862.x
Verbruggen E, Veresoglou SD, Anderson IC, Caruso T, Hammer EC, Kohler J et al (2013) Arbuscular mycorrhizal fungi - short-term liability but long-term benefits for soil carbon storage? New Phytol 197:366–368. https://doi.org/10.1111/nph.12079
Veresoglou SD, Chen B, Rillig MC (2012) Arbuscular mycorrhiza and soil nitrogen cycling. Soil Biol Biochem 46:53–62. https://doi.org/10.1016/J.SOILBIO.2011.11.018
Vestergård M, Henry F, Rangel-Castro JI, Michelsen A, Prosser JI, Christensen S (2008) Rhizosphere bacterial community composition responds to arbuscular mycorrhiza, but not to reductions in microbial activity induced by foliar cutting. FEMS Microbiol Ecol 64:78–89. https://doi.org/10.1111/j.1574-6941.2008.00447.x
Walker AP, De Kauwe MG, Bastos A, Belmecheri S, Georgiou K, Keeling R et al (2020). Integrating the evidence for a terrestrial carbon sink caused by increasing atmospheric CO 2. New Phytol nph.16866. https://doi.org/10.1111/nph.16866.
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Wang H, Boutton TW, Xu W, Hu G, Jiang P, Bai E et al (2015) Quality of fresh organic matter affects priming of soil organic matter and substrate utilization patterns of microbes. Sci Rep 5:10102. https://doi.org/10.1038/srep10102
Weisser WW, Roscher C, Meyer ST, Ebeling A, Luo G, Allan E et al (2017) Biodiversity effects on ecosystem functioning in a 15-year grassland experiment: Patterns, mechanisms, and open questions. Basic Appl Ecol 23:1–73. https://doi.org/10.1016/j.baae.2017.06.002
Whiteside MD, Werner GDA, Caldas VEA, van’t Padje A, Dupin SE, Elbers B et al. (2019). Mycorrhizal Fungi Respond to Resource Inequality by Moving Phosphorus from Rich to Poor Patches across Networks. Curr Biol 29, 2043-2050.e8. https://doi.org/10.1016/J.CUB.2019.04.061.
Wickham H (2011) ggplot2. Wiley Interdiscip Rev Comput Stat 3:180–185. https://doi.org/10.1002/wics.147
Williams A, Manoharan L, Rosenstock NP, Olsson PA, Hedlund K (2017) Long-term agricultural fertilization alters arbuscular mycorrhizal fungal community composition and barley ( Hordeum vulgare ) mycorrhizal carbon and phosphorus exchange. New Phytol 213:874–885. https://doi.org/10.1111/nph.14196
Xu X, Thornton PE, Post WM (2013) A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob Ecol Biogeogr 22:737–749. https://doi.org/10.1111/geb.12029
Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi S et al (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480. https://doi.org/10.1038/s41564-018-0129-3
Zhang J, Kobert K, Flouri T, Stamatakis A (2014) PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. https://doi.org/10.1093/bioinformatics/btt593
Zhu Q, Riley WJ, Tang J (2017) A new theory of plant-microbe nutrient competition resolves inconsistencies between observations and model predictions. Ecol Appl 27:875–886. https://doi.org/10.1002/eap.1490
Acknowledgements
SC received funding from long term DAAD scholarship to carry out the research. ML is funded by the German Research Foundation (DFG; FOR 456, FOR 1451 – “The Jena Experiment”) and by the “Zwillenberg-Tietz Stiftung”. We acknowledge help from Agnes Fastnacht with greenhouse resources and monitoring of the experiment. Special thanks to Karl Kübler for construction and deployment of the pulse labelling setup in the greenhouse. We acknowledge Heike Geilmann and Steffen Ruehlow for help with stable isotope measurements, and Maria Foerster for helping with fatty acid analysis. We also thank Erika Kothe, Ruchira Mukherji, Elisa Catao and Huei Ying Gan for helpful comments and discussions and Simon Benk for proof reading the MS.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
SC, GG and AM designed the study. SC and JH performed the experiment. SC performed the extractions and stable isotope analyses. TG and RG completed the downstream processing of nucleic acids, sequencing and supervised bioinformatics analysis. SC and ML completed the statistical analysis. SC, ML and GG conceptualized the work and wrote the manuscript and all authors contributed to revisions.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Responsible Editor: Tatsuhiro Ezawa.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chowdhury, S., Lange, M., Malik, A.A. et al. Plants with arbuscular mycorrhizal fungi efficiently acquire Nitrogen from substrate additions by shaping the decomposer community composition and their net plant carbon demand. Plant Soil 475, 473–490 (2022). https://doi.org/10.1007/s11104-022-05380-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05380-x