Abstract
Background and aims
Phosphorus (P) is an essential nutrient for plant growth, but is also one of the least accessible in soil. Plants have evolved several strategies to cope with P deficiency and recently the role of the phytohormones strigolactones (SLs) in modulating tomato plants acclimation to P shortage has been described. How SLs regulate the use of P from sparingly accessible P sources, such as organic P or precipitated metal-P systems, is however still unknown in tomato.
Methods
In this study, we compared P acquisition strategies of wild-type (WT) and SL-depleted tomato plants grown hydroponically in the presence of dissolved inorganic phosphate (Pi), dissolved myo-inositol hexaphosphate (myoInsP6), or their coprecipitated form following Fe(II) oxidative precipitation.
Results
Irrespective of the P treatment, SL-depleted plants accumulated more P in their tissues than the WT, possibly due to the constitutively higher expression of high-affinity P transporters and activity of P-hydrolyzing enzymes. Wild-type plants were conversely more effective at acidifying their growth medium and exuding more organic compounds in the presence of dissolved myoInsP6 or coprecipitated forms of P, but this behaviour did not translate into a higher P acquisition.
Conclusions
The two genotypes activated different subsets of responses to bypass low P bioavailability, although the P acquisition efficiency (PAE) was not effectively increased. Strigolactone-depleted plants achieved higher PAE values than WT plants regardless of the applied P form, highlighting a central role of SLs in controlling P uptake and optimizing the cost/benefit ratio of P acquisition.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Phosphorus (P) is one of the essential mineral nutrients required by plants for their growth and balanced metabolism. However, due to its extremely low bioavailability in soil, P is often limiting for crop productivity in over 40% of arable land (Vance 2001). The slow diffusion coefficient of inorganic P (Pi) in the form of orthophosphate ion (10−12 - 10−15 m2 s−1, Rausch and Bucher 2002) and its strong interaction with soil constituents result in a P concentration in soil solution ranging from 0.1 to 10 μM, which is inadequate to support plant growth, metabolism and production (Frossard et al. 2000; Raghothama 1999; Zhang et al. 2011). In particular, a variety of soil processes such as the high P adsorption on soil minerals, especially iron (Fe) and aluminium (Al) oxides, P complexation by calcium (Ca), Al or Fe to form metal-P salts (Akhtar et al. 2008; Vance et al. 2003) and oxidative coprecipitation of P with Fe(II) (Santoro et al. 2019), all contribute to making P one of the least mobile macronutrients in many natural and agricultural ecosystems. In addition, the organic P pool represents a potentially important source for plant nutrition accounting for 20–80% of the total P in soil (Hayes et al. 2000). This pool is mainly composed of phosphomonoesters, as inositol hexaphosphates, which however require enzymatic hydrolysis prior to acquisition by plants, and can undergo the same soil retention mechanisms as Pi (Celi et al. 2020; Dalal 1978; Santoro et al. 2019).
Increasing evidence ascertains that some plant species efficiently utilize certain sparingly accessible P sources, determining morphological, physiological and biochemical modifications of the rhizosphere (Thennegedara and Dissanayaka 2021; Wang et al. 2011; Zhang et al. 2011; Zhou et al. 2021). These include adjustments of root traits and architecture such as length, surface area, fineness, and root hair density (Rao et al. 1999), rhizosphere acidification through the release of protons (H+), root exudation of organic acid anions, upregulation of genes encoding high-affinity P transporters (PHOSPHATE TRANSPORTER, PHT), enhanced production and secretion of P-hydrolyzing enzymes, and symbiotic associations with mycorrhizal fungi (Adu-Gyamfi et al. 2009; Akhtar et al. 2008; Jin et al. 2014; Wang and Lambers 2020; Zhang et al. 2011; Zhou et al. 2021).
Rhizosphere acidification that occurs when P availability is low can play an important role in the mobilization of phosphate in neutral and alkaline calcareous soils (Akhtar et al. 2008; Zhang et al. 2011), as the efflux of H+ to soil through ATPase pumps located in the plasma membrane can lead to a significant decrease in soil pH, particularly in poorly buffered soils, favouring the dissolution of sparingly soluble P phases (Akhtar et al. 2008). Furthermore, root exudation of carboxylates, which could also contribute to rhizosphere acidification, is an effective strategy to increase P availability through complexation (Akhtar et al. 2008; Zhang et al. 2011). Indeed, carboxylates may have a greater potential than phosphate to chelate cations in the soil matrix, thereby releasing P into solution (Gerke et al. 2000a, b; Jones 1998; Ryan et al. 2001). In some species, the composition and concentration of carboxylates in root exudates varies according to the P sources in the soil (Lambers et al. 2002). Finally, the release by the roots of extracellular enzymes such as phosphatases in response to P deficiency has been implicated in the improvement of plant P nutrition (Chen et al. 2002). Similarly, phytases that specifically hydrolyze metal (Fe, Al, Ca) phytates can be released. Such phytate salts constitute an important fraction of the total organic P present in the soil (Turner et al. 2002). Phytases also represent a way to recycle plant internal P pools, since phytates are also a major P storage form in many plant tissues (Konietzny and Greiner 2002). Internal plant P homeostasis is systemically regulated by the conserved microRNA 399 (miR399), which acts as a long-distance signal highly responsive to low P conditions (Czarnecki et al. 2013), and is thought to be a crucial node of the P starvation response (PSR) (Pant et al. 2008). miR399 targets the transcript of PHOSPHATE2 (PHO2), encoding an ubiquitin conjugating enzyme that mediates the degradation of Phosphate Starvation-Induced (PSI) gene products, like PHO1 and PHT family member proteins (Liu et al. 2012).
Recent studies have indicated a major role of strigolactones (SLs), a group of carotenoid-derived phytohormones, as signalling molecules able to trigger morphological, physiological and biochemical responses associated with plant acclimation to P deficient conditions (Czarnecki et al. 2013; Ito et al. 2015; Gamir et al. 2020; Marro et al. 2021). Strigolactone biosynthesis and exudation have been shown to increase under P starvation in many species such as rice, Arabidopsis and tomato (Niu et al. 2013). In this regard, Marro et al. (2021) recently proposed that the induced SL biosynthesis under P deficiency would modulate the expression of key P signalling and regulatory genes, and that of P transporters. In particular, SLs were proposed to act on the SPX-PHR1 complex releasing the transcriptional activator PHR1, thus inducing the increase of mature miR399 and the expression of TPSI1, a non-protein coding gene involved in miR399 sequestration (Marro et al. 2021). In turn, miR399 reduces the transcript levels of the PSR suppressor PHO2, activating the PSR pathway with the induced expression of PHT transporters-encoding genes and consequent P acquisition (Gamir et al. 2020; Marro et al. 2021). The involvement of SLs in the regulation of plant responses to P deficiency likely occurs through their crosstalk with other phytohormones, such as cytokinins, auxin and ethylene, thus highlighting a complex network of hormonal signals involved in response to P scarcity (Kapulnik et al. 2011; Villaécija-Aguilar et al. 2019). Recently, we have reported that SLs partly mediate several physiological and morphological changes triggered by P deficiency (Santoro et al. 2020, 2021). We have observed that SL-depleted tomato plants differ from wild-type (WT) controls at the level of root morphology, as many root traits were more severely affected at low P provision (80 μM) (Santoro et al. 2020). This condition induced many P-deficiency responses in SL-depleted plants, which WT plants only adopted under harsh P stress conditions, including extensive cell and tissue disorganization at the root tip (Santoro et al. 2020, 2021). The enhanced P uptake, expression of PHT genes and root enzymatic activity observed in SL-depleted plants compared to WT plants may be related to an altered perception of exogenous P and/or of endogenous P levels. On the other hand, some responses such as root acidification in response to low P were initially delayed and impaired in the absence of SLs (Santoro et al. 2021). The role of SLs in the ability of tomato plants to access different forms of sparingly available P sources, such as dissolved organic P or precipitated metal-P systems, is however still largely unknown.
In this study, we aimed at evaluating differences in the response between WT and SL-depleted tomato plants in mining P from sparingly available organic and inorganic P forms. We hypothesized that tomato plants would react to the presence of these P forms by triggering the PSR at least in part – both within root tissues and in terms of exudate composition, and that this process may be affected by SLs. To this purpose, we assessed plant growth and elemental composition, assayed the activity of root P-related enzymes and the expression of PHT genes, and characterized root exudates of WT and SL-depleted tomato plants grown hydroponically in the presence of dissolved Pi, dissolved myo-inositol hexaphosphate (myoInsP6, or phytate) and the coprecipitated form of these two P-containing compounds following Fe(II) oxidative precipitation.
Materials and methods
Plant material and growth conditions
The tomato (Solanum lycopersicum L.) SlCCD7-silenced line 6936 (SL-depleted) and its WT genotype M82 assayed in this study were kindly provided by Dr. H.J. Klee (University of Florida); in the former, the production of the major SLs is reduced by about 80–90% with respect to the latter (Vogel et al. 2010). Seeds were surface sterilized in 70% (v/v) ethanol for 30 s, then in 15% NaClO for 20 min, washed five times for 5 min with sterile water, and then germinated on wet Whatman filter paper in Petri dishes (10 cm diameter) at 25 °C and in darkness for 5 days. Germinated seeds were transferred to plastic pots filled with P-free silica sand and allowed to grow for 45 days in a growth chamber with a 16/8 h light/dark cycle, air temperature of 25 °C and relative humidity ≥70%, with a light intensity of 100 μmol m−2 s−1. Pots were irrigated daily with a Hoagland modified nutrient solution containing the following salts: 1 mM MgSO4, 1 mM Ca(NO3)2, 250 μM KNO3, 80 μM KH2PO4, 20 μM FeNaEDTA, 9 μM H3BO3, 1.8 μM MnCl2, 0.2 μM ZnSO4, 0.2 μM Co(NO3)2, 0.2 μM NiSO4, 0.2 μM CuSO4 (pH adjusted to 6.0).
Phosphorus nutritional conditions and collection of root exudates
After 45 days of growth in sand, plants were transplanted to 250 mL flasks containing 200 mL of aerated nutrient solution. After two days of acclimation to the hydroponic growth systems with the complete nutrient solution, roots were gently rinsed with deionized water to remove traces of P. After that, four plants per genotype were kept for 15 days in a P-replete nutrient solution (Pi, 80 μM KH2PO4), in a P-free (–P, 0 μM KH2PO4) nutrient solution, or in a nutrient solution with P provided as the organic form myoInsP6, at a final P concentration of 80 μM. In addition to these conditions, four plants per genotype were supplied with sparingly available P sources in the form of Fe-P coprecipitates with Pi or myoInsP6 (referred to as COP-Pi and COP-myoInsP6, respectively), synthesized as described by Santoro et al. (2019) and having the chemical properties reported in Table 1. The amount of each coprecipitate added to the nutrient solution was chosen in order to obtain a final P concentration of 80 μM. In all treatments (except Pi) KCl replaced KH2PO4 to provide plants with a steady amount of K. The solutions/suspensions were constantly aerated during the experiment and were replenished every day in order to restore the initial volume. After 15 days, root exudates were collected, filtered through 0.22 μm nylon membrane filters and stored at −20 °C for further analyses. Plants were harvested, divided into shoots and roots, and the fresh biomass was recorded. Root subsamples were frozen in liquid N2 and stored at −80 °C for enzymatic and molecular analyses, while the remaining root and shoot tissues were dried at +40 °C, ground separately in a mortar, passed through a 0.5 mm mesh sieve and used for elemental analyses.
Plant elemental analysis
Concentration of total P in plant tissues was determined colorimetrically on dry plant material after sulfuric-perchloric digestion using the malachite green method (Ohno and Zibilske 1991). Phosphorus absorption efficiency (PAE) values were calculated as the ratio of P accumulated in tissues to P exogenously supplied during both plant growth in sand and hydroponics (Neto et al. 2016). After acidic dissolution, Fe concentration in roots was measured by atomic absorption spectrometry (PerkinElmer AAnalyst 1400, Norwalk, CT, USA). Total C and N contents were determined by dry combustion (UNICUBE, Elementar Analysensysteme GmbH, Langensenbold, Germany).
Exudate analysis
Root exudates were analyzed for dissolved organic C (DOC), protons, organic acid anions, total P, and Fe content. DOC was determined using Pt-catalyzed, high-temperature combustion (850 °C) followed by infrared detection of CO2 (VarioTOC, Elementar, Hanau, Germany), after removing inorganic C by acidifying to pH 2 and purging with CO2-free synthetic air. A blank to correct for C derived from EDTA in the nutrient solution was performed. In addition, UV absorption at λ = 254 nm was measured (Helios Gamma Spectrophotometer, Thermo Electron, Waltham, MA). The specific UV absorbance (SUVA) values, calculated by normalizing measured absorbance values to the concentration of DOC, were used as an estimate of the aromatic content of exudates samples (Weishaar et al. 2003).
The concentration of protons in the nutrient solutions was monitored using a pH-sensitive electrode (inoLab pH 7110, WTW GmbH, Weilheim, Germany) and was expressed as the mmoles of released H+ ions per grams of root DW. Organic acid anions were analyzed by Dionex DX-500 Ion Chromatography system (Sunnyvale, CA, USA) equipped with a dimensional-exclusion column (Ion PAC ICE-AS6) and an electrochemical detector (Dionex ED40). The concentration of P in the nutrient solution was determined colorimetrically as described above, while Fe concentration in the solution was measured by atomic absorption spectrometry (PerkinElmer AAnalyst 1400, Norwalk, CT, USA).
Dissolved organic C and organic acid anions data were expressed as mg C g−1 plant DW to evaluate the exudates in terms of C investment by the plant.
Enzymatic activity
Phosphatase and phytase activities were determined as described in Hayes et al. (1999). Root material was ground in 15 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.5) containing 0.5 mM CaCl2·H2O and 1 mM EDTA. The extract was centrifuged at 13,800 g × 15 min at 4 °C and the supernatant gel-filtered at 4 °C on Sephadex G-25 columns. To assay total acid phosphatase activity, the enzyme extract was incubated at 26 °C in 15 mM MES buffer (pH 5.5) with 1 mM EDTA, 5 mM cysteine and 10 mM p-nitrophenyl phosphate (pNPP). The reaction was stopped after 30 min by addition of 0.25 M NaOH. The concentration of p-nitrophenol (pNP) was determined by measuring the absorbance at 412 nm against standard solutions. Phytase activity was measured on the same root extracts and under the same conditions described above, except that pNPP was replaced with 2 mM potassium myoInsP6. The reaction was stopped after 60 min by addition of ice-cold 10% trichloroacetic acid (TCA), and Pi concentration was determined by the malachite green method (Ohno and Zibilske 1991).
Gene transcript quantification
To quantify PHO2 and PHT gene transcripts, RNA was extracted from individual root samples of WT and SL-depleted plants grown in hydroponics according to the conditions described above. Total RNA was extracted by using Spectrum™ Plant Total RNA Kit (Sigma-Aldrich), and treated with DNase I (ThermoScientific) at 37 °C for 30 min to remove residual genomic DNA. First-strand cDNA was synthesized from 500 ng of purified total RNA using the High-Capacity cDNA Reverse-Transcription Kit (Applied Biosystems, Monza, Italy) according to the manufacturer’s instructions. The specific primer pairs are the same as reported in Santoro et al. (2021). The quantitative reverse-transcriptase PCR (qRT-PCR) reactions were carried out in a StepOnePlus system (Applied Biosystems) using the SYBR Green (Applied Biosystems) method. Transcript concentrations were normalized on SlEF-1α transcripts as endogenous controls and quantified through the 2-ΔΔCt method. Four independent biological replicates were analyzed, and each qRT-PCR reaction was run in technical triplicates.
Statistics
For all determinations, the analysis of variance (one-way ANOVA) was performed using the SPSS software version 27.0 (SPSS, Chicago, IL, USA), and was followed by pair-wise post-hoc analyses (Student-Newman-Keuls test) to determine which means differed significantly at p < 0.05. The different P treatments were firstly compared for each genotype separately, then a pairwise comparison of the two genotypes subjected to the same P treatment was carried out. In addition, to evaluate the effect of plant genotype, P treatment and the combination of the two on the measured parameters, we used a linear mixed-effect ANOVA model performed with the statistical programming language R (R Core Team 2020). Figures were created using SigmaPlot ver. 12.5 software (Systat, San Jose, CA, USA) showing means presented with standard errors.
Results
Plant growth parameters and elemental composition
Strigolactone-depleted genotypes typically displayed increased shoot branching in comparison to WT plants (not shown). The root biomass was highly dependent on the P treatment (p < 0.001), the plant genotype (p < 0.01) and the combination of the two (p < 0.05). In particular, both genotypes achieved the maximal and comparable dry root biomass under the Pi condition (Table 2). Similarly, the root biomass did not differ between genotypes when they were starved for P. Minimum values of root biomass were associated with plants grown on coprecipitates, even though the roots of SL-depleted plants were generally more developed compared to the WT, especially when they received COP-Pi (Table 2).
Similar to the roots, shoot growth was strongly influenced by the P treatment (p < 0.001), the plant genotype (p < 0.001) and the combination of the two (p < 0.001). The highest shoot biomass was attained by SL-depleted plants under Pi and myoInsP6 conditions (Table 2). Also, under –P and in the presence of COP-Pi SL-depleted plants was higher than that of the WT. For both genotypes, the lowest shoot biomass was observed in the presence of COP-myoInsP6 (Table 2).
The R/S ratios of SL-depleted plants were lower than in the WT genotype when P was supplied as myoInsP6 or under –P conditions, due to the higher shoot biomass (Table 2). The R/S ratio of WT plants was generally lower in the presence of coprecipitates with respect to the dissolved forms, while the opposite was observed for SL-depleted plants. Also in this case, both P treatment (p < 0.01) and the plant genotype (p < 0.05), as well as the combination of the two (p < 0.001), concurred to influence the R/S ratio. The P uptake in roots and shoots is reported in Fig. 1a, while the total P uptake per plant is reported in Fig. 1b. Total P uptake was generally higher in both genotypes treated with dissolved P forms with respect to insoluble forms (Fig. 1b). In all cases, the shoot P content was higher in SL-depleted plants than in the WT (Fig. 1a), reflecting the greater shoot biomass production by the mutants. In roots, P contents followed the same trend as in the shoots in response to the different treatments, but they were almost comparable between the two genotypes (Fig. 1a). For both genotypes, the minimum accumulation of P in the plant was observed in the presence of COP-myoInsP6, with a higher value in SL-depleted plants, which was however comparable to –P conditions (Fig. 1b), consistently with differences in P content at the shoot level (Fig. 1a). Root P content was influenced mainly by the P treatment (p < 0.001) and to a lesser extent by the plant genotype (p < 0.05), which conversely played a more significant role in the accumulation of P in the shoots (p < 0.001), along with the combination of the two parameters (p < 0.001). The greater P uptake by SL-depleted plants led to generally higher P-acquisition efficiency (PAE) than the WT, for the same amount and form of P provided (Fig. 1c). The maximal PAE value was shown by SL-depleted plants under Pi conditions. Similarly, when supplied with the coprecipitates and dissolved myoInsP6, SL-depleted plants displayed higher PAE than the WT (Fig. 1c). In general, PAE values were more strongly influenced by the plant genotype (p < 0.001) than by the P treatment (p < 0.05), and plants grown in the presence of the two coprecipitates displayed the lowest PAE among all P treatments (Fig. 1c).
a) Root and shoot P content, b) total plant P content and c) P-acquisition efficiency (PAE) of wild-type (WT) and SL-depleted (SL–) tomato plants after 15 days of hydroponic culture without P (–P) or with different P forms (Pi, myoInsP6, COP-Pi, COP-myoInsP6). Each value represents the mean of four biological replicates (± SE). Different letters above bars indicate significant differences (p < 0.05) when all treatments were compared for each genotype separately. Asterisks indicate significant differences resulting from the pairwise comparison of the two genotypes subjected to the same P treatment (p < 0.05)
The contents of C, N and Fe in plants are reported in Table 3. In general, SL-depleted plants displayed higher C contents than the WT, especially when grown with myoInsP6 or COP-Pi. The lowest C and N contents were found in plants supplied with COP-myoInsP6, irrespective of the genotype (Table 3). Nitrogen values differed among treatments, with SL-depleted plants generally accumulating more N than WT plants. N/P ratios showed a clear increase when Pi and myoInsP6 were provided as coprecipitates or plants were grown without P (Table 3). Both C and N contents in plants were significantly influenced by the P treatment (p < 0.001) and the plant genotype (p < 0.001), but not by their combined effect.
The highest Fe content in root tissues was found in plants provided with either COP-Pi or COP-myoInsP6, without statistical differences between genotypes (Table 3). The lowest Fe accumulation was recorded under Pi, with WT plants displaying significantly lower values than SL-depleted plants, and myoInsP6 conditions (Table 3). Iron accumulation in roots was significantly influenced by the P treatment (p < 0.001) and less by the plant genotype (p < 0.05). In contrast, it was not affected by the combined effect of the P treatment and the plant genotype.
Root enzymatic activity
Phosphatase activity was maximal in WT and SL-depleted plants grown in the presence of COP-myoInsP6 or COP-Pi, without statistical differences between genotypes, while minimum in Pi and –P plants (Fig. 2a). The provision of P in the form of dissolved myoInsP6 increased the activity of phosphatases compared to Pi condition in both genotypes (Fig. 2a). The trend of phytase activity was similar to that described for phosphatase, with a significantly higher activity in SL-depleted plants under Pi, myoInsP6 or COP-Pi conditions (Fig. 2b). Both enzyme activities were significantly influenced by the P treatment (p < 0.0001) and, to a lesser extent, by the plant genotype (p < 0.05).
a) Phosphatase and b) phytase activity in roots of wild-type (WT) and SL-depleted (SL–) tomato plants after 15 days of hydroponic growth without P (–P) or with different P forms (Pi, myoInsP6, COP-Pi, COP-myoInsP6). Each value represents the mean of four biological replicates (± SE). Different letters above bars indicate significant differences (p < 0.05) when all treatments were compared for each genotype separately. Asterisks indicate significant differences resulting from the pairwise comparison of the two genotypes subjected to the same P treatment (p < 0.05)
Expression of genes encoding high-affinity P transporters
The gene encoding PHOSPHATE TRANSPORTER 1 (LePT1) was up-regulated in SL-depleted plants compared to the WT, regardless of the P treatment applied (Fig. 3a). In the WT, LePT1 showed similar transcript concentration in plants supplied with dissolved or sparingly available P sources, but higher in plants under –P than in Pi. LePT2 was most expressed under –P condition in both genotypes, with the maximum reported for SL-depleted plants (Fig. 3b). Its lowest transcript concentration was found in plants under Pi and plants subjected to the COP-Pi treatment. The trend of LePT4 in the WT genotype was similar to that of LePT2, while it was the highest in SL-depleted plants in the presence of COP-Pi (Fig. 3c). Finally, in our experiment, PHO2 expression in WT plants was highest when supplied with readily available Pi (Fig. 3d). In SL-depleted plants, PHO2 was abundantly expressed when P was provided as coprecipitate (COP-Pi or COP-myoInsP6) or myoInsP6, while a lower expression than in the WT was evident in plants under Pi (confirming the findings in Santoro et al. 2021).
Relative transcript concentrations of the phosphate transporter-encoding genes a) LePT1, b) LePT2, c) LePT4, and of d) SlPHO2 in roots of wild-type (WT) and SL-depleted (SL–) tomato plants after 15 days of hydroponic growth without P (–P) or with different P forms (Pi, myoInsP6, COP-Pi, COP-myoInsP6). Each value represents the mean of four biological replicates (± SE) normalized over SlEF-1α transcripts and quantified through the 2-ΔΔCt method. Different letters above bars indicate significant differences (p < 0.05) when all treatments were compared for each genotype separately. Asterisks indicate significant differences resulting from the pairwise comparison of the two genotypes subjected to the same P treatment (p < 0.05)
Exudate analysis
At the end of the 15 days of the experiment, no dissolved P or Fe were detected in the solutions of either WT or SL-depleted plants under any P treatment, while part of the coprecipitate was still present in the nutrient solutions of plants treated with COP-Pi and COP-myoInsP6.
Dissolved organic C exudation
The amount of C exuded tended to be higher in WT than in SL-depleted plants, and the treatment with COP-myoInsP6 was the most effective in triggering the exudation in both genotypes (Fig. 4). Furthermore, C exudation was greater in plants grown under P starvation compared to Pi plants, although differences were only significant for WT plants. When P was provided as dissolved myoInsP6 or COP-Pi, C exudation was comparable within genotypes. The lowest C exudation was recorded for SL-depleted plants with Pi or COP-Pi (Fig. 4).
Dissolved organic C (DOC, bars) content and respective SUVA values (white circles) in the exudates of wild-type (WT) and SL-depleted (SL–) tomato plants after 15 days of hydroponic growth without P (–P) or with different P forms (Pi, myoInsP6, COP-Pi, COP-myoInsP6). Each value represents the mean of four replicates (± SE) expressed as mg of C per gram of plant dry weight. Different letters above (DOC) or within bars (SUVA) indicate significant differences (p < 0.05) when all treatments were compared for each genotype separately. Asterisks indicate significant differences resulting from the pairwise comparison of the two genotypes subjected to the same P treatment (p < 0.05)
The high C exudation by plants grown with COP-myoInsP6 was also accompanied by relatively high SUVA values (circles in Fig. 4), while fairly high SUVA values were additionally observed when P was provided as dissolved myoInsP6. The lowest SUVA levels were determined in root exudates of SL-depleted plants grown on COP-Pi, Pi and under –P conditions. Finally, the statistical analysis revealed that C exudation was significantly influenced by both the P treatment (p < 0.001) and the plant genotype (p < 0.001), while the aromaticity of the exuded compounds was only affected by the P treatment (p < 0.001).
Proton exudation
Proton release by WT and SL-depleted plants was significantly dependent on the P treatment only (p < 0.001). The highest amount of exuded protons was observed in WT plants grown in the presence of myoInsP6, while the lowest proton release was observed in plants grown under –P, Pi, and COP-Pi conditions (Fig. 5). Slightly larger amounts of protons were instead exuded by both genotypes in the presence of COP-myoInsP6 (Fig. 5).
H+ content of exudates of wild-type (WT) and SL-depleted (SL–) tomato plants after 15 days of hydroponic growth without P (–P) or with different P forms (Pi, myoInsP6, COP-Pi, COP-myoInsP6). Each value represents the mean of four biological replicates (± SE) expressed as mmoles of H+ per gram of root dry weight. Different letters above bars indicate significant differences (p < 0.05) when all treatments were compared for each genotype separately. Asterisks indicate significant differences resulting from the pairwise comparison of the two genotypes subjected to the same P treatment (p < 0.05)
Organic acid anion exudation
Quantitative analysis of organic anions revealed variable amounts of oxalic, succinic, tartaric, and citric acid anions, while others were only detectable in traces. Oxalate was the dominant organic anion in root exudates, and its concentration increased in –P plants (Fig. 6a). In the presence of COP-Pi and COP-myoInsP6, oxalate exudation from WT plants showed an increasing, although not significant, trend when compared to the relative soluble form, while it slightly decreased in SL-depleted plants grown on COP-Pi. On the contrary, succinate exudation was not triggered by the presence of COP-Pi, while it barely increased in both genotypes under COP-myoInsP6 with respect to dissolved myoInsP6, and under –P conditions in WT plants only (Fig. 6b). The exudation of tartrate by WT plants was maximal under Pi conditions and not detectable when plants were grown with myoInsP6 or COP-myoInsP6 (Fig. 6c). In SL-depleted plants, while again undetectable on myoInsP6 and COP-myoInsP6, tartrate in the exudates accumulated more under P-deficiency and low P bioavailability. Conversely, and in contrast with WT plants, it was low in the presence of Pi (Fig. 7c). Citrate exudation by both genotypes significantly increased when plants received COP-Pi or were P starved, reaching significantly higher values in WT than SL-depleted plants under –P conditions, while it was not detected in the presence of dissolved or coprecipitated myoInsP6, for either genotype (Fig. 6d).
Average concentration of identified organic acid anions in the root exudates of wild-type (WT) and SL-depleted (SL–) tomato plants after 15 days of hydroponic growth without Pi (–P) or with different Pi forms (Pi, myoInsP6, COP-Pi, COP-myoInsP6). Each value represents the mean of four biological replicates (± SE) expressed as mg of C per gram of plant dry weight. Different letters above bars indicate significant differences (p < 0.05) when all treatments were compared for each genotype separately. Asterisks indicate significant differences resulting from the pairwise comparison of the two genotypes subjected to the same P treatment (p < 0.05)
Discussion
Plant growth and P acquisition by different P forms are mediated by SLs
So far, neither the ability of tomato plants to absorb various P pools with different availability, nor the role of SLs in the underlying processes have been reported in the literature. Our data indicated that both the WT and SL-depleted genotypes are able to use P from myoInsP6 when provided as a dissolved molecule, but less so when coprecipitated with Fe. Furthermore, P acquisition by plants grown with COP-myoInsP6 is lower compared to plants supplied with COP-Pi. Consistent with these findings, Findenegg and Nelemans (1993) have found that Zea mays plants grown in quartz sand can use myoInsP6 as a source of P for nutrition, but their growth is severely reduced when myoInsP6 is added to a soil with high P-retaining capacity. Similarly, myoInsP6 has been reported to be equivalent to Pi as a P source for Lupinus spp. in sand, but represents a poorer P source in soil (Adams and Pate 1992). Martin et al. (2004) also observed limited availability of myoInsP6 to Lolium perenne L. when adsorbed on goethite. It should be noted, however, that not all plants are able to use myoInsP6 as a source of P. Hayes et al. (2000), for instance, reported that six pasture species could obtain little P from soluble myoInsP6 when grown in sterile media, even when myoInsP6 was supplied at high concentrations (up to 40 times the level of Pi supply required for plants to reach 90% of their maximum attainable shoot dry weight). Species-dependent differences in P acquisition from myoInsP6 could be due to a diverse root phytase activity associated with PSRs. In support of this hypothesis, tomato plants, especially when SL-depleted, produced significantly more phytases in the presence of myoInsP6 than under Pi conditions, which could explain the elevated P acquisition from dissolved myoInsP6. High phytase activity was also determined in WT and SL-depleted plants supplied with COP-myoInsP6, albeit with little effect on P acquisition. Indeed, it has been previously reported that phytase activity is inhibited when myoInsP6 is retained by soil surfaces (George et al. 2007). In addition, Giaveno et al. (2010) have pointed out that phytases are not able to hydrolyze this organic P compound when adsorbed on various iron oxides. Thus, we can infer that also Fe(II) oxidative coprecipitation could hamper myoInsP6 hydrolysis by phytases and limit P acquisition by plants.
Irrespective of the P treatment applied, SL-depleted plants produced a greater shoot biomass and accumulated more P than the WT, confirming the different phenotypic traits between these two genotypes reported by Santoro et al. (2021). Together with the greater activity of P-hydrolyzing enzymes, the higher P uptake by SL-depleted plants could be the result of an increased expression of high-affinity P transporters at the Pi values used in this work. With respect to PHT genes expression, our results only in part confirm those reported by Gamir et al. (2020) and Marro et al. (2021). Discrepancy in some outcomes (e.g., expression of LePT2) could be due to the different P concentrations applied to plants: 80 vs 800–1300 μM Pi for P-sufficient conditions, and 0 vs 200–300 μM Pi for the P depleted conditions in our work and in those of Gamir et al. (2020) and Marro et al. (2021), respectively. We adopted the 80 μM Pi concentration to mimic the range plants are likely subjected to in the field (Hinsinger 2001). This concentration revealed sufficient for the WT but could uncover a defect in SL-depleted plants, likely due to altered perception and/or production of internal P stocks: it would be worth investigating whether this defect may be directly linked to the production or perception of the signalling molecule InsP8, the inositol pyrophosphate that is directly perceived by the endogenous sensing system, switching the PSR off (Riemer et al. 2021). Alternatively, or in parallel, SL-defective plants may be less efficient in P utilization, which may keep feedback repression of PSR responses off; the miR399-PHO module is indeed dysregulated in SL-deficient plants at 80 μM Pi (Santoro et al. 2021). It is thus possible that this defective trait is conditional to sub-limiting Pi availability, and as such is not apparent at higher Pi concentrations as reported earlier (Gamir et al. 2020; Marro et al. 2021), where a less intense PSR was registered in SL-deficient plants. On the contrary, in our experiments, the transcription of LePT1, LePT2, and LePT4 was higher in SL-depleted roots under –P and/or Pi conditions with respect to the WT. PHO2 transcripts were instead decreased under Pi conditions in these plants, in line with the typical P starvation response (Bari et al. 2006; Santoro et al. 2021). PHO2 is in fact an ubiquitin-conjugating E2-ligase that in Arabidopsis is needed to degrade the PHO1 protein (Liu et al. 2012). A close homologue has been identified in tomato (Zhao et al. 2019). PHO1 in turn is critical to P translocation to shoots; unsurprisingly, PHO2 expression is decreased during the P starvation response so that pho2 mutants over-accumulate P in the shoot, while pho1 mutants do so in the roots (Delhaize and Randall 1995; Hamburger et al. 2002; Zhao et al. 2019). When different P forms than Pi were supplied to WT and SL-depleted plants, the picture was however more nuanced and depended on the PHT gene. LePT1 transcripts were higher in SL-depleted plants irrespective of the P supply form, and for LePT2 in the absence of P or with Pi, but not so for the other conditions. The enhanced PHT transcription, together with phytase activity, may justify why SL-depleted plants tended to be more effective (higher PAE values) at accessing not only Pi and myoInsP6 but also sparingly soluble P (COP-Pi or COP-myoInsP6) than WT plants. It is interesting to note that PHO2 transcripts correlated inversely with P uptake and especially shoot P content of WT and SL-depleted plants under Pi and –P conditions (see also Santoro et al. 2021), but that such correlation was lost when P is supplied in other forms than Pi. A comparison with shoot P contents suggests that the transcriptional regulation of PHO2 may not be a key factor in the translocation of P when supplied in different forms than Pi, both in WT and SL-depleted plants. It would be worth to investigate the pattern of P allocation in PHO1-edited plants (Zhao et al. 2019) fed with myoInsP6 or coprecipitates, in order to evaluate whether the P over-accumulation in the shoot of SL-depleted plants is lost in the absence of PHO1 gene functions. To our knowledge indeed, despite all the information gathered so far, the role of the PHO2-PHO1 module in P translocation when P is supplied under different forms than Pi is not known, in any plant species.
The treatment of plants with soluble myoInsP6 was associated to a high N uptake in both genotypes. We hypothesize that the higher N accumulation, together with the negative correlation between P and N concentration in plants (ρ = −0.821, p < 0.001) and in line with previous results reported by Santoro et al. (2021), could be related to the larger synthesis of exoenzymes that operate P mineralization (Marklein and Houlton 2012). In fact, although N concentration in plants usually decreases with increasing P limitation (de Groot et al. 2003), several studies have shown that adding N to P-starved plants enhances phosphatase activity, which in turn indicates that plants use N to mobilize and acquire P from organic sources and to increase the internal recycling of P, reduce the short-term P deficits and therefore delay the onset of P limitation symptoms (Heuck et al. 2018; Marklein and Houlton, 2021; Schleuss et al. 2020; Widdig et al. 2019). The content of N in both genotypes grown on coprecipitates or under –P was however lower than in the presence of Pi or soluble myoInsP6. Nevertheless, as pointed out by Koerselman and Meuleman (1996), the ratio between N and P provides a better indication of whether N or P limit plant growth, rather than the absolute content of these nutrients in plant tissue. Thus, the nature of nutrient limitation can be directly established from the N/P ratio in plant tissues, with ratios higher than 16 indicating P limitation, whereas ratios lower than 14 suggest N limitation (Koerselman and Meuleman 1996). Under our experimental conditions, the N/P ratios of both WT and SL-depleted plants were higher than 16 regardless of the P source applied. However, the N/P ratios significantly decreased with Pi and myoInsP6 if compared to the –P treatment, suggesting that P limitation was alleviated. On the contrary, plants grown on COP-Pi and COP-myoInsP6 displayed higher N/P ratios, despite the lower N content in tissues, as –P control plants. Furthermore, the N/P ratio under these treatments (COP-Pi, COP-myoInsP6, –P) was lower in SL-depleted than WT plants. These ratios could result from the higher P acquisition by SL-depleted plants and the generally higher expression of PHT transporters and activity of P-hydrolyzing enzymes when SL biosynthesis is reduced.
The sparingly available forms of P disfavoured the allocation of dry matter to the roots in WT plants, resulting in R/S ratios even lower than those observed under Pi conditions, while the same ratio remained unchanged in SL-depleted plants. It is known that some plant species allocate more C to the root system in P-deficient soils resulting in increased R/S ratio and greater exploration of the surface soil, where sparingly available P forms are mainly accumulated (Rao et al. 2016; Vance et al. 2003). However, Edayilam et al. (2018) have observed that Andropogon virginicus grown in the presence of FePO4 or Ca3(PO4)2 relies more on physiological modifications for P foraging rather than on root morphology alterations, while other plant species experiencing P deficiency, such as Lupinus albus, L. cosentinii and Cicer arietinum, are reported to depend on both mechanisms, by enhancing P acquisition through increased root exudation following the formation of cluster-roots (Pearse et al. 2007). We have recently observed (Santoro et al. 2020) that when WT tomato plants are grown under low P conditions (10 μM Pi) their total root length and tip number are increased compared to plants supplied with higher P levels (80 μM Pi) showing that this species relies on both physiological and morphological acclimation mechanisms. We have also demonstrated that SLs take part in the orchestration of this multifaceted response, since SL-depleted plants show no variations and even a reduction of these same root parameters when moved from low P to no P supply. In the current study, when the low P availability was due to the application of coprecipitates to plants, the physiological responses probably dominated over the morphological changes and this could explain why we did not observe any significant increase in the R/S ratio.
Root exudation in response to different P forms is modulated by SLs
Regardless of the applied P treatment, tomato plants tended to acidify their growth medium. The amount of protons in root exudates negatively correlated with the plant biomass (ρ = −0.621, p < 0.05) and P content in the plant (ρ = −0.668, p < 0.05), indicating that high levels of protons in the exudates were likely due to an increased need for P uptake. In both genotypes, the addition of myoInsP6 triggered a sharp increase in proton release by roots, but a moderate increase when provided as coprecipitate. Possibly, both genotypes increased the release of protons to favour the hydrolysis of myoInsP6 by phytases, that show an optimal pH close to 5 (Giaveno et al. 2010). In addition, the same hydrolytic reaction can produce protons, therefore contributing to the solution acidification and justifying the high acidity of the exudates.
When SL-depleted plants were supplied with myoInsP6 their growth was substantially the same as with Pi, while in the case of WT plants we noticed a decrease in biomass in spite of (and possibly, to some extent, because of) the more protons and DOC exuded. These findings suggest that SLs might be involved in limiting P uptake from organic soluble forms such as myoInsP6 in order to optimize the cost/benefit ratio of P acquisition. Interestingly, we observed a net decline in biomass production and P accumulation when plants of both genotypes were supplied with myoInsP6 coprecipitated with Fe. We postulate that the significant increase in proton exudation triggered in the presence of COP-myoInsP6 may have caused a remarkable oxide dissolution, thus increasing the concentration of both P and Fe in the solution. This might explain the high Fe concentration in the roots of WT and SL-depleted plants treated with COP-myoInsP6. Nevertheless, we cannot exclude that the increase of Fe uptake is a consequence of P starvation (Rouached et al. 2010). Also, given the negative correlation of root Fe concentration with the root biomass (ρ = −0.662, p < 0.05), root growth inhibition may be caused by Fe toxicity at the root tip (Rouached et al. 2010; Ward et al. 2008) due to a high level of reactive oxygen species, resulting in increased callose deposition in cell walls and plasmodesmata (Müller et al. 2015). The enhanced callose deposition, probably triggered by redox signalling started by the ferroxidase LPR1 (LOW PHOSPHATE ROOT1), might interfere with the intercellular movement of the SHORT ROOT protein, finally impairing root growth. Therefore, the accumulation of Fe to toxic levels in the roots of plants treated with COP-myoInsP6 or COP-Pi could be a possible cause of their limited growth, also considering that their biomass and capacity to accumulate P were lower compared to –P plants.
With respect to plants under Pi and –P conditions, those supplied with COP-Pi did not differ in the amount of protons exuded per gram of root, suggesting that proton extrusion was not the principal strategy activated to retrieve P from Fe-P coprecipitates. The amount of DOC exuded from plants fed with COP-Pi was higher than from Pi-treated plants, consistently with a greater exudation of some organic acid anions. This was particularly evident for WT plants, whereas exudation of C increased only slightly in SL-depleted plants treated with COP-Pi.
In agreement with previous results (Santoro et al. 2021), SL-depleted plants exuded the lowest amount of C when grown with Pi, while under –P conditions their C exudation increased. The profile of organic anions confirmed that the majority of C exuded consisted of oxalate, while other anions such as succinate, tartrate, and citrate contributed less. For most organic anions, the exudation was triggered more by the absence of P than by the presence of a sparingly available P form compared to the plants supplied with dissolved P. The addition of myoInsP6 to plants led to higher levels of C exudation if compared to Pi, especially by WT plants. According to the SUVA values, the C-bearing compounds in the exudates from SL-depleted plants treated with COP-myoInsP6 were the most aromatic, so they possibly included molecules, such as polyphenols, that could promote the release of P by reductive dissolution of the oxide to which it is bound (Juszczuk et al. 2004). Thus, considering the highest values of DOC exudation and SUVA index found for plants grown with COP-myoInsP6, we could infer that this coprecipitate may have been subjected to a greater dissolution by aromatic molecules in addition to the action of P-hydrolyzing enzymes in the SL-depleted genotype.
In general, the two genotypes preferentially activated different subsets of responses to bypass P shortage or low P bioavailability. Although these strategies did not lead to higher PAE than Pi-supplied plants, they did allow SL-depleted plants to achieve higher PAE values than WT plants when fed with coprecipitates. In general, SL-depleted plants tended to express PHT genes more than the WT under comparable conditions, with a few exceptions. Despite the relatively acidic pH, the elevated amount of DOC exuded and the enhanced activity of P-hydrolyzing enzymes in roots, neither genotypes were able to acquire P from coprecipitates, resulting in growth traits that were substantially comparable to those observed in P-deprived plants. One possible reason is that these strategies may have negatively affected each other. Proton release into a poorly buffered nutrient solution could lower the pH, shifting the carboxylic acid/carboxylate ratio towards the acid form, and therefore reducing their efficacy in displacing P from the coprecipitate or dissolving the oxide, at least in the case of COP-myoInsP6 (Pearse et al. 2007). In addition, Zhao and Wu (2014) have reported that the maximum P-extracting capability from a calcareous soil by four plant species (two woody Moraceae and two herbaceous cruciferous plants) occurred after 40 days of observation, concurrently with the maximal C losses, and Edayilam et al. (2018) observed differences in P uptake from poorly available forms of P (Ca3(PO4)2 and FePO4) after 16 weeks of growth. It is thus possible that the duration of the experiment in our study did not allow for an efficient activation of mechanisms that scavenge the nutrient from coprecipitates. Furthermore, it should be noted that the properties of the sparingly available P sources play a pivotal role in determining the extent of P absorption by plants. The coprecipitates used in this experiment had similar mesopore volume, but quite different surface charge (+2 mV COP-Pi vs − 38 mV COP-myoInsP6). The elevated and negative surface charge of COP-myoInsP6 could have prevented organic acid anions to approach the coprecipitate surface due to electrostatic repulsion, resulting in decreased efficiency of ligand-exchange/dissolution mechanisms. Conversely, COP-Pi surface is positively charged and more enriched in P than COP-myoInsP6, allowing organic acids to compete with P for retention sites.
Conclusions
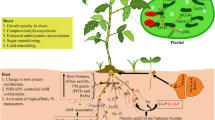
In this study, SL-depleted plants produced more shoot biomass and accumulated more P than the WT, regardless of the P form applied. This was likely justified by the increased expression of P transporters and activity of P-hydrolyzing enzymes, which appear to be constitutive traits of the SL-depleted genotype, possibly resulting from the altered perception and/or metabolism of internal P stocks. Both genotypes could use P from dissolved myoInsP6, due to the greater activity of P-hydrolyzing enzymes and proton exudation, but were less efficient in using P from myoInsP6 when it was coprecipitated with Fe, perhaps because of the enzymatic inhibition stemmed from the retention of the molecule by a solid phase. However, the C-bearing compounds in the exudates of SL-depleted plants treated with COP-myoInsP6 were the most aromatic, so they conceivably included molecules, such as polyphenols, that could potentially promote the release of P by reductive dissolution of the oxide. In plants supplied with COP-Pi, C exudation rather than proton extrusion was the main strategy activated to retrieve P. As a general conclusion, we can state that although the different strategies activated by the two genotypes in the presence of sparingly accessible P forms did not lead to higher PAE than Pi plants, they did allow SL-depleted plants to achieve higher PAE values than WT plants, confirming the central role of SLs in controlling P uptake and balancing resource costs for P acquisition (as summarized in Fig. 7). Further studies could better focus on the molecular and biochemical mechanisms that underline the differences in P acquisition from different sparingly available P forms and in its translocation within the plants using more available mutants of the model species A. thaliana.
Conclusive conceptual summary. 1) Both wild-type (WT) and SL-depleted (SL–) plants could use P from soluble myoInsP6, while 2) they were not able to retrieve P from coprecipitates. 3a) OA anion exudation was triggered by P deficiency rather than the presence of COP-Pi, possibly because of a delayed activation of responses to sparingly available P. 3b) Both myoInsP6 forms induced similar responses in the two genotypes, indicating a prevailing influence of the P form over SL control of the PSR, apart from the constitutively higher expression of P transporters in SL– plants. 4) However, under either P condition, P uptake and accumulation was higher in SL– plants, highlighting the central role of these hormones in optimal P management. Abbreviations: SL, strigolactone; COP, coprecipitate; DOC, dissolved organic C; myoInsP6, myo-inositol hexaphosphate; OA, organic acid; PSR, phosphate-starvation response
Abbreviations
- Pi:
-
Inorganic phosphate
- myoInsP6:
-
myo-inositol hexaphosphate
- SLs:
-
strigolactones
- WT:
-
Wild-type
- COP:
-
Coprecipitate
References
Adams MA, Pate JS (1992) Availability of organic and inorganic forms of phosphorus to lupins (Lupinus spp.). Plant Soil 145:107–113. https://doi.org/10.1007/BF00009546
Adu-Gyamfi JJ, Aigner M, Gludovacz D (2009) Variations in phosphorus acquisition from sparingly soluble forms by maize and soybean in low-and medium-P soils using P-32. Proc Int Plant Nutr Colloquium XVI:1008–1016
Akhtar MS, Oki Y, Adachi T (2008) Genetic variability in phosphorus acquisition and utilization efficiency from sparingly soluble P-sources by Brassica cultivars under P-stress environment. J Agron Crop Sci 194:380–392. https://doi.org/10.1111/j.1439-037X.2008.00326.x
Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 14(3):988–999. https://doi.org/10.1104/pp.106.079707
Celi L, Prati M, Magnacca G, Santoro V, Martin M (2020) Role of crystalline iron oxides on stabilization of inositol phosphates in soil. Geoderma 374:114442. https://doi.org/10.1016/j.geoderma.2020.114442
Chen CR, Condron LM, Davis MR, Sherlock RR (2002) Phosphorus dynamics in the rhizosphere of perennial ryegrass (Lolium perenne L.) and radiata pine (Pinus radiata D. Don.). Soil Biol Biochem 34(4):487–499. https://doi.org/10.1016/S0038-0717(01)00207-3
Czarnecki O, Yang J, Weston DJ, Tuskan GA, Chen JG (2013) A dual role of strigolactones in phosphate acquisition and utilization in plants. Int J Mol Sci 14:7681–7701. https://doi.org/10.3390/ijms14047681
Dalal RC (1978) Organic phosphorus. Adv Agron 29:83–117
Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107(1):207–213. https://doi.org/10.1104/pp.107.1.207
de Groot CC, Marcelis LFM, van den Boogaard R, Kaiser WM, Lambers H (2003) Interaction of nitrogen and phosphorus nutrition in determining growth. Plant Soil 248:257–268. https://doi.org/10.1023/A:1022323215010
Edayilam N, Montgomery D, Ferguson B, Maroli AS, Martinez N, Powell BA, Tharayil N (2018) Phosphorus stress-induced changes in plant root exudation could potentially facilitate uranium mobilization from stable mineral forms. Environ Sci Technol 52:7652–7662. https://doi.org/10.1021/acs.est.7b05836
Findenegg GR, Nelemans JA (1993) The effect of phytase on the availability of P from myo-inositol hexaphosphate (phytate) for maize roots. Plant Soil 154:189–196. https://doi.org/10.1007/BF00012524
Frossard E, Condron LM, Oberson A, Sinaj S, Fardeau JC (2000) Processes governing phosphorus availability in temperate soils. J Environ Qual 29:12–53
Gamir J, Torres-Vera R, Rial C, Berrio E, de Souza Campos PM, Varela RM, Macías FA, Pozo MJ, Flors V, López-Ráez JA (2020) Exogenous strigolactones impact metabolic profiles and phosphate starvation signalling in roots. Plant Cell Environ 43:1655–1668. https://doi.org/10.1111/pce.13760
George TS, Simpson RJ, Hadobas PA, Marshall DJ, Richardson AE (2007) Accumulation and phosphatase-lability of organic phosphorus in fertilised pasture soils. Aust J Agric Res 58(1):47–55. https://doi.org/10.1071/AR06167
Gerke J, Beissner L, Römer W (2000) The quantitative effect of chemical phosphate mobilization by carboxylate anions on P uptake by a single root. I The basic concept and determination of soil parameters J Plant Nutr Soil Sci 163:207–212
Gerke J, Beissner L, Römer W (2000) The quantitative effect of chemical phosphate mobilization by carboxylate anions on P uptake by a single root. II The importance of soil and plant parameters for uptake of mobilized P J Plant Nutr Soil Sci 163:213–219
Giaveno C, Celi L, Richardson AE, Simpson RJ, Barberis E (2010) Interaction of phytases with minerals and availability of substrate affect the hydrolysis of inositol phosphates. Soil Biol Biochem 42(3):491–498. https://doi.org/10.1016/j.soilbio.2009.12.002
Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14(4):889–902. https://doi.org/10.1105/tpc.000745
Hayes JE, Richardson AE, Simpson RJ (1999) Phytase and acid phosphatase activities in extracts from roots of temperate pasture grass and legume seedlings. Aust J Plant Physiol 26(8):801–809. https://doi.org/10.1071/PP99065
Hayes JE, Richardson AE, Simpson RJ (2000) The growth and phosphorus utilization of plants in sterile media when supplied with inositol hexaphosphate, glucose 1-phosphate or inorganic phosphate. Plant Soil 220:165–174. https://doi.org/10.1023/A:1004782324030
Hinsinger P (2001) Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237:173–195. https://doi.org/10.1023/A:1013351617532
Heuck C, Smolka G, Whalen ED, Frey S, Gundersen P, Moldan F, Fernandez IJ, Spohn M (2018) Effects of long-term nitrogen addition on phosphorus cycling in organic soil horizons of temperate forests. Biogeochemistry 141:167–181. https://doi.org/10.1007/s10533-018-0511-5
Ito S, Nozoye T, Sasaki E, Imai M, Shiwa Y, Shibata-Hatta M, Ishige T, Fukui K, Ito K, Nakanishi H, Nishizawa NK, Yajima S, Asami T (2015) Strigolactone regulates anthocyanin accumulation, acid phosphatases production and plant growth under low phosphate condition in Arabidopsis. PLoS One 10(3):e0119724. https://doi.org/10.1371/journal.pone.0119724
Jin J, Tang C, Hogarth TW, Armstrong R, Sale P (2014) Nitrogen form but not elevated CO2 alters plant phosphorus acquisition from sparingly soluble phosphorus sources. Plant Soil 374:109–119. https://doi.org/10.1007/s11104-013-1870-2
Jones DL (1998) Organic acids in the rhizosphere – a critical review. Plant Soil 205:25–44. https://doi.org/10.1023/A:1004356007312
Juszczuk IM, Wiktorowska A, Malusa E, Rychter AM (2004) Changes in the concentration of phenolic compounds and exudation induced by phosphate deficiency in bean plants (Phaseolus vulgaris L.). Plant Soil 267:41–49. https://doi.org/10.1007/s11104-005-2569-9
Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H (2011) Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J Exp Bot 62:2915–2924. https://doi.org/10.1093/jxb/erq464
Koerselman W, Meuleman AF (1996) The vegetation N: P ratio: a new tool to detect the nature of nutrient limitation. J Appl Ecol 33:1441–1450. https://doi.org/10.2307/2404783
Konietzny U, Greiner R (2002) Molecular and catalytic properties of phytate-degrading enzymes (phytases). Int J Food Sci Technol 37:791–812. https://doi.org/10.1046/j.1365-2621.2002.00617.x
Lambers H, Juniper D, Cawthray GR, Veneklaas EJ, Martínez-Ferri E (2002) The pattern of carboxylate exudation in Banksia grandis (Proteaceae) is affected by the form of phosphate added to the soil. Plant Soil 238:111–122. https://doi.org/10.1023/A:1014289121672
Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WI, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24(5):2168–2183. https://doi.org/10.1105/tpc.112.096636
Marklein AR, Houlton BZ (2012) Nitrogen inputs accelerate phosphorus cycling rates across a wide variety of terrestrial ecosystems. New Phytol 193:696–704. https://doi.org/10.1111/j.1469-8137.2011.03967.x
Marro N, Lidoy J, Chico MÁ, Rial C, García J, Varela RM, Mácias FA, Pozo MJ, Janoušková M, López-Ráez JA (2021) Strigolactones: new players in the nitrogen-phosphorus signalling interplay. Plant Cell Environ:1–16. https://doi.org/10.1111/pce.14212
Martin M, Celi L, Barberis E (2004) Desorption and plant availability of myo-inositol hexaphosphate adsorbed on goethite. Soil Sci 169(2):115–124. https://doi.org/10.1097/01.ss.0000117787.98510.9d
Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S (2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell 33:26–230. https://doi.org/10.1016/j.devcel.2015.02.007
Neto AP, Favarin JL, Hammond JP, Tezotto T, Couto HTZ (2016) Analysis of phosphorus use efficiency traits in coffea genotypes reveals Coffea arabica and Coffea canephora have contrasting phosphorus uptake and utilization efficiencies. Front Plant Sci 7:408. https://doi.org/10.3389/fpls.2016.00408
Niu YF, Chai RS, Jin GL, Wang H, Tang CX, Zhang YS (2013) Responses of root architecture development to low phosphorus availability: a review. Ann Bot 112:391–408. https://doi.org/10.1093/aob/mcs285
Ohno T, Zibilske LM (1991) Determination of low concentration of phosphorus in soil extracts using malachite green. Soil Sci Soc Am J 55(3):892–895. https://doi.org/10.2136/sssaj1991.03615995005500030046x
Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53:731–738. https://doi.org/10.1111/j.1365-313X.2007.03363.x
Pearse SJ, Veneklaas EJ, Cawthray G, Bolland MDA, Lambers H (2007) Carboxylate composition of root exudates does not relate consistently to a crop species’ ability to use phosphorus from aluminium, iron or calcium phosphate sources. New Phytol 173:181–190. https://doi.org/10.1111/j.1469-8137.2006.01897.x
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. Accessed 1 June 2021
Raghothama KG (1999) Phosphorus and plant nutrition: An overview. In: Sims JT, Sharpley AN (eds) Phosphorus: Agriculture and the environment. ASA, CSSA, and SSSA, Madison, WI, pp 355–378. https://doi.org/10.2134/agronmonogr46.c11
Rao IM, Friesen DK, Osaki M (1999) Plant adaptation to phosphorus-limited tropical soils. In: Pessarakli M (ed) Handbook of plant and crop stress. Marcel Dekker, New York, pp 61–96
Rao IM, Miles JW, Beebe SE, Horst WJ (2016) Root adaptations to soils with low fertility and aluminium toxicity. Ann Bot 118(4):593–605. https://doi.org/10.1093/aob/mcw073
Rausch C, Bucher M (2002) Molecular mechanisms of phosphate transport in plants. Planta 216:23–37. https://doi.org/10.1007/s00425-002-0921-3
Riemer E, Qiu D, Laha D, Harmel RK, Gaugler P, Gaugler V, Frei M, Hajirezaei MR, Laha NP, Krusenbaum L, Schneider R, Saiardi A, Fiedler D, Jessen HJ, Schaaf G, Giehl RFH (2021) ITPK1 is an InsP6/ADP phosphotransferase that controls phosphate signaling in Arabidopsis. Mol Plant 14(11):1864–1880. https://doi.org/10.1016/j.molp.2021.07.011
Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3:288–299. https://doi.org/10.1093/mp/ssp120
Ryan PR, Delhaize E, Jones DL (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52:527–560. https://doi.org/10.1146/annurev.arplant.52.1.527
Santoro V, Martin M, Persson P, Lerda C, Said-Pullicino D, Magnacca G, Celi L (2019) Inorganic and organic P retention by coprecipitation during ferrous iron oxidation. Geoderma 348:168–180. https://doi.org/10.1016/j.geoderma.2019.04.004
Santoro V, Schiavon M, Gresta F, Ertani A, Cardinale F, Sturrock CJ, Celi L, Schubert A (2020) Strigolactones control root system architecture and tip anatomy in Solanum lycopersicum L. plants under P starvation. Plants 9:612. https://doi.org/10.3390/plants9050612
Santoro V, Schiavon M, Visentin I, Constán-Aguilar C, Cardinale F, Celi L (2021) Strigolactones affect phosphorus acquisition strategies in tomato plants. Plant Cell Environ 44(11):3628–3642. https://doi.org/10.1111/pce.14169
Schleuss PM, Widdig M, Heintz-Buschart A, Kirkman K, Spohn M (2020) Interactions of nitrogen and phosphorus cycling promote P acquisition and explain synergistic plant-growth responses. Ecology 101(5):e03003. https://doi.org/10.1002/ecy.3003
Thennegedara TGRM, Dissanayaka DMSB (2021) Phosphorus mobilizing capacity of selected grain legumes grown under phosphorus-deficient conditions. Trop Agric Res 32(4):380–389. https://doi.org/10.4038/tar.v32i4.8413
Turner BL, McKelvie ID, Haygarth PM (2002) Characterisation of water extractable soil organic phosphorus by phosphatase hydrolysis. Soil Biol Biochem 34:27–35. https://doi.org/10.1016/S0038-0717(01)00144-4
Vance CP (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127(2):390–397. https://doi.org/10.1104/pp.010331
Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157:423–447. https://doi.org/10.1046/j.1469-8137.2003.00695.x
Villaécija-Aguilar JA, Hamon-Josse M, Carbonnel S, Kretschmar A, Schmid C, Dawid C, Gutjahr C (2019) SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signaling in Arabidopsis. PLoS Genet 15(8):e1008327. https://doi.org/10.1371/journal.pgen.1008327
Vogel JT, Walter MH, Giavalisco P, Lytovchenko A, Kohlen W, Charnikhova T, Simkin AJ, Goulet C, Strack D, Bouwmeester HJ, Fernie AR, Klee HJ (2010) SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J 61:300–311. https://doi.org/10.1111/j.1365-313X.2009.04056.x
Wang X, Guppy CN, Watson L, Sale PWG, Tang C (2011) Availability of sparingly soluble phosphorus sources to cotton (Gossypium hirsutum L.), wheat (Triticum aestivum L.) and white lupin (Lupinus albus L.) with different forms of nitrogen as evaluated by a 32P isotopic dilution technique. Plant Soil 348:85–98. https://doi.org/10.1007/s11104-011-0901-0
Wang Y, Lambers H (2020) Root-released organic anions in response to low phosphorus availability: recent progress, challenges and future perspectives. Plant Soil 447:135–156. https://doi.org/10.1007/s11104-019-03972-8
Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147:1181–1191. https://doi.org/10.1104/pp.108.118562
Weishaar JL, Aiken GR, Bergamaschi BA, Fram MS, Fujii R, Mopper K (2003) Evaluation of specific ultraviolet absorbance as an indicator of the chemical composition and reactivity of dissolved organic carbon. Environ Sci Technol 37(20):4702–4708. https://doi.org/10.1021/es030360x
Widdig M, Schleuss PM, Weig AR, Guhr A, Biederman LA, Borer ET, Crawley MJ, Kirkman KP, Seabloom E, Wragg P (2019) Nitrogen and phosphorus additions alter the abundance of phosphorus-solubilizing bacteria and phosphatase activity in grassland soils. Front Environ Sci 7:185. https://doi.org/10.3389/fenvs.2019.00185
Zhang HW, Huang Y, Ye XS, Xu FS (2011) Genotypic variation in phosphorus acquisition from sparingly soluble P sources is related to root morphology and root exudates in Brassica napus. Sci China Life Sci 54:1134–1142. https://doi.org/10.1007/s11427-011-4254-y
Zhao K, Wu Y (2014) Rhizosphere calcareous soil P-extraction at the expense of organic carbon from root-exuded organic acids induced by phosphorus deficiency in several plant species. Soil Sci Plant Nutr 60:640–650. https://doi.org/10.1080/00380768.2014.934191
Zhao P, You Q, Lei M (2019) A CRISPR/Cas9 deletion into the phosphate transporter SlPHO1;1 reveals its role in phosphate nutrition of tomato seedlings. Physiol Plant 167(4):556–563. https://doi.org/10.1111/ppl.1289
Zhou W, Chen D, Zeng Q, Tahir MA, Wu Q, Huang Y, Jiang Y, Li Q, Ao J, Huang Z (2021) Differential physiological behavior of sugarcane genotypes in response to sparingly soluble phosphorus-sources. J Plant Nutr Soil Sci 184:187–197. https://doi.org/10.1002/jpln.202000333
Acknowledgments
We would like to show our gratitude to Federica Matera for technical assistance.
Funding
This research has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 727929 (A novel and integrated approach to increase multiple and combined stress tolerance in plants using tomato as a model - TOMRES).
Author information
Authors and Affiliations
Contributions
Conceptualization: Veronica Santoro, Michela Schiavon, Luisella Celi; Methodology: Veronica Santoro, Michela Schiavon, Luisella Celi; Formal analysis and investigation: Veronica Santoro, Ivan Visentin, Maria Martin, Daniel Said-Pullicino; Writing - original draft preparation: Veronica Santoro, Michela Schiavon; Writing - review and editing: all authors; Funding acquisition: Francesca Cardinale, Luisella Celi; Supervision: Luisella Celi. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Additional information
Responsible Editor: Honghua He.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Santoro, V., Schiavon, M., Visentin, I. et al. Tomato plant responses induced by sparingly available inorganic and organic phosphorus forms are modulated by strigolactones. Plant Soil 474, 355–372 (2022). https://doi.org/10.1007/s11104-022-05337-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-022-05337-0