Abstract
Aims
Soil microbiome roles in agriculture is becoming more and more important. This importance is also reflected on the way plants are seen: complex organisms formed by the plant itself plus the microbes inhabiting its tissues, including the ones on the surface of every organ and the ones adhered or in proximity to the roots. In addition, as already demonstrated, the microbial community associated with a specific soil is able to predetermine the health status of crops. For all the above mentioned reasons, defining the microbial composition of agricultural soils and the factors driving the assemblage is pivotal to achieve more sustainable agriculture and viticulture.
Methods
We aimed to investigate how the soil geological characteristics influence the microbiome composition associated with close geographically related vineyards. Moreover, we studied both the top (15 cm in depth) and deep (120 cm in depth) soil layers as anthropically influenced and almost-undisturbed soil, respectively.
Results
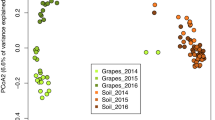
We observed slightly different microbial communities despite the close geographical proximity of the two vineyards, which is considered one of the main determinants of the soil microbiome composition. In addition, we found that the geological characteristics of the two soils influence both the root distribution and the accumulation of pathogen- and symbiont-related genera. Sensory profiles of the Grillo wines from the two different soils confirmed the tight link between soil origin and wine traits.
Conclusions
In the present study, we highlight that the geological characteristics of soil can influence soil microbial composition and assemblage in close geographically related vineyards, with a potential effect on wine features.






Similar content being viewed by others
Data availability
The SRA accession numbers of the NGS reported in this paper are deposited in NCBI under the BioProject PRJNA655455; BioSample SAMN15735114 and SAMN15735115; SRA accession SRR12436974 and SRR12436973.
Code availability
Not applicable.
References
Abarenkov K, Henrik Nilsson R, Larsson KH, Alexander IJ, Eberhardt U, Erland S et al (2010) The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol 186:281–285
Agustí-Brisach C, Mostert L, Armengol J (2014) Detection and quantification of Ilyonectria spp. associated with black‐foot disease of grapevine in nursery soils using multiplex nested PCR and quantitative PCR. Plant Pathol 63:316–322
Alabi OJ, Casassa LF, Gutha LR, Larsen RC, Henick-Kling T, Harbertson JF, Naidu RA (2016) Impacts of grapevine leafroll disease on fruit yield and grape and wine chemistry in a wine grape (Vitis vinifera L.) cultivar. PLoS One 11:e0149666
Alagna F, Balestrini R, Chitarra W, Marsico A, Nerva L (2020) Getting ready with the priming: Innovative weapons against biotic and abiotic crop enemies in a global changing scenario. In: Priming-Mediated Stress and Cross-Stress Tolerance in Crop Plants. Elsevier, Amsterdam, pp 35–56
Angel R (2012) Total nucleic acid extraction from soil. Protoc Exch 10
Aroca Á, Gramaje D, Armengol J, García-Jiménez J, Raposo R (2010) Evaluation of the grapevine nursery propagation process as a source of Phaeoacremonium spp. and Phaeomoniella chlamydospora and occurrence of trunk disease pathogens in rootstock mother vines in Spain. Eur J Plant Pathol 126:165–174
Arroyo-García R, Ruiz‐Garcia L, Bolling L, Ocete R, Lopez M, Arnold C et al (2006) Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol Ecol 15:3707–3714
Balestrini R, Lumini E (2018) Focus on mycorrhizal symbioses. Appl Soil Ecol 123:299–304
Balestrini R, Chitarra W, Fotopoulos V, Ruocco M (2017) Potential role of beneficial soil microorganisms in plant tolerance to abiotic stress factors. In: Soil Biological Communities and Ecosystem Resilience. Springer, Berlin, pp 191–207
Balestrini R, Chitarra W, Antoniou C, Ruocco M, Fotopoulos V (2018) Improvement of plant performance under water deficit with the employment of biological and chemical priming agents. J Agric Sci 156:680–688
Berdeja M, Nicolas P, Kappel C, Dai ZW, Hilbert G, Peccoux A et al (2015) Water limitation and rootstock genotype interact to alter grape berry metabolism through transcriptome reprogramming. Hortic Res 2:15012
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Böhm W (1979) Profile wall methods. Methods of Studying Root Systems. Springer, Berlin, pp 48–60
Böhm W, Köpke U (1977) Comparative root investigations with two profile wall methods [Oats]. Zeitschrift fuer Acker-und Pflanzenbau (Germany, FR).
Bokulich NA, Thorngate JH, Richardson PM, Mills DA (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci 111: E139-E148
Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F et al (2012) Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91
Cabrala A, Rego C, Crous PW, Oliveira H (2012) Virulence and cross-infection potential of Ilyonectria spp. to grapevine. Phytopathol Mediterranea, 340–354
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581
Cameron DD, Neal AL, van Wees SC, Ton J (2013) Mycorrhiza-induced resistance: more than the sum of its parts? Trends Plant Sci 18:539–545
Capece A, Romaniello R, Siesto G, Pietrafesa R, Massari C, Poeta C, Romano P (2010) Selection of indigenous Saccharomyces cerevisiae strains for Nero d’Avola wine and evaluation of selected starter implantation in pilot fermentation. Int J Food Microbiol 144:187–192
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335
Chitarra W, Perrone I, Avanzato CG, Minio A, Boccacci P, Santini D et al (2017) Grapevine grafting: Scion transcript profiling and defense-related metabolites induced by rootstocks. Front Plant Sci 8:654
Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y et al (2013) Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642
Coller E, Cestaro A, Zanzotti R, Bertoldi D, Pindo M, Larger S et al (2019) Microbiome of vineyard soils is shaped by geography and management. Microbiome 7:140
Fendrihan S, Constantinescu F, Sicuia O-A, Dinu S (2016) Beneficial Bacillus strains improve plant resistance to phytopathogens: a review. Int J Environ Agric Biotechnol 1(2): 238512
Fierer N (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579
Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC (2009) Global patterns in belowground communities. Ecol Lett 12:1238–1249
Fileccia V, Ruisi P, Ingraffia R, Giambalvo D, Frenda AS, Martinelli F (2017) Arbuscular mycorrhizal symbiosis mitigates the negative effects of salinity on durum wheat. PLoS One 12.9: e0184158
Gayevskiy V, Goddard MR (2012) Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J 6:1281
Gilbert JA, van der Lelie D, Zarraonaindia I (2014) Microbial terroir for wine grapes. Proc Natl Acad Sci 111:5–6
Giménez-Jaime A, Aroca A, Raposo R, García‐Jiménez J, Armengol J (2006) Occurrence of fungal pathogens associated with grapevine nurseries and the decline of young vines in Spain. J Phytopathol 154:598–602
Gramaje D, Armengol J (2011) Fungal trunk pathogens in the grapevine propagation process: potential inoculum sources, detection, identification, and management strategies. Plant Dis 95:1040–1055
Granett J, Goheen A, Lider L, White J (1987) Evaluation of grape rootstocks for resistance to type A and type B grape phylloxera. Am J Enol Vitic 38:298–300
Hammer Ø, Harper D, Ryan P (2001) PAST-Palaeontological statistics. http://www.uves/~pardomv/pe/2001_1/past/pastprog/past.pdf. Accessed on June 2020
Harshavardhan M, Kumar P (2018) Putrescine and glomus mycorrhiza mitigate salinity induced stress responses in sorghum (SSV-74). In: Lovely Professional University, Phagwara
Jung SC, Martinez-Medina A, Lopez-Raez JA, Pozo MJ (2012) Mycorrhiza-induced resistance and priming of plant defenses. J Chem Ecol 38:651–664
Knight S, Klaere S, Fedrizzi B, Goddard MR (2015) Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Sci Rep 5:14233
Kolton M, Erlacher A, Berg G, Cytryn E (2016) The Flavobacterium genus in the plant holobiont: ecological, physiological, and applicative insights. In: Microbial models: From environmental to industrial sustainability. Springer, Berlin, pp 189–207
Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R (2012) Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 13:47
Kwak M-J, Kong HG, Choi K, Kwon S-K, Song JY, Lee J et al (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 36:1100–1109
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R et al (2013) Fungal community analysis by high-throughput sequencing of amplified markers–a user’s guide. New Phytol 199:288–299
Liu D, Chen Q, Zhang P, Chen D, Howell K (2020) Vineyard ecosystems are structured and distinguished by fungal communities impacting the flavour and quality of wine. BioRxiv: 2019.2012. 2027.881656
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Manici L, Saccà M, Caputo F, Zanzotto A, Gardiman M, Fila G (2017) Long-term grapevine cultivation and agro-environment affect rhizosphere microbiome rather than plant age. Appl Soil Ecol 119:214–225
Mannino G, Nerva L, Gritli T, Novero M, Fiorilli V, Bacem M et al (2020) Effects of different microbial inocula on tomato tolerance to Water Deficit. Agronomy 10:170
Marasco R, Rolli E, Fusi M, Michoud G, Daffonchio D (2018) Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6:3
McMurdie PJ, Holmes S (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531
Mezzasalma V, Sandionigi A, Guzzetti L, Galimberti A, Grando MS, Tardaguila J, Labra M (2018) Geographical and cultivar features differentiate grape microbiota in Northern Italy and Spain vineyards. Front Microbiol 9:946
Miura T, Sánchez R, Castañeda LE, Godoy K, Barbosa O (2017) Is microbial terroir related to geographic distance between vineyards? Environ Microbiol Rep 9:742–749
Nayarisseri A, Suppahia A, Nadh AG, Nair AS (2015) Identification and characterization of a pesticide degrading flavobacterium species EMBS0145 by 16S rRNA gene sequencing. Interdisciplinary Sciences: Computational Life Sciences 7:93–99
Nerva L, Zanzotto A, Gardiman M, Gaiotti F, Chitarra W (2019) Soil microbiome analysis in an ESCA diseased vineyard. Soil Biol Biochem 135:60–70
Parte SG, Mohekar AD, Kharat AS (2017) Microbial degradation of pesticide: a review. Afr J Microbiol Res 11:992–1012
Pelsy F (2010) Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity 104:331
Pennock D, McKenzie N, Montanarella L (2015) Status of the world’s soil resources. Technical Summary FAO, Rome
Pozo MJ, Azcón-Aguilar C (2007) Unraveling mycorrhiza-induced resistance. Curr Opin Plant Biol 10:393–398
Pozo MJ, Jung SC, Martínez-Medina A, López-Ráez JA, Azcón-Aguilar C, Barea J-M (2013) Root allies: arbuscular mycorrhizal fungi help plants to cope with biotic stresses. In: Symbiotic endophytes. Springer, Berlin, pp 289–307
Pretorius IS (2000) Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729
Proffitt T, Campbell-Clause J (2012) Managing grapevine nutrition and vineyard soil health. Wines of Western Australia. Grape Wine Research Development Corporation (GWRDC), Claremont, 32
Raaijmakers JM, Mazzola M (2016) Soil immune responses. Science 352:1392–1393
Rivers AR, Weber KC, Gardner TG, Liu S, Armstrong SD (2018) ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 7
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864
Scienza A, Giorgianni A (2015) Atlante geologico dei vini d’Italia: vitigno, suolo e fattori climatici. Giunti Editore, Florence, pp 204–211
Serna-Chavez HM, Fierer N, Van Bodegom PM (2013) Global drivers and patterns of microbial abundance in soil. Glob Ecol Biogeogr 22:1162–1172
Shafi J, Tian H, Ji M (2017) Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotechnol Equip 31:446–459
Stirling G, Cirami R (1984) Resistance and tolerance of grape rootstocks to South Australian populations of root-knot nematode. Aust J Exp Agric 24:277–282
Tristezza M, Fantastico L, Vetrano C, Bleve G, Corallo D, Grieco F, Mita G (2014) Molecular and technological characterization of Saccharomyces cerevisiae strains isolated from natural fermentation of Susumaniello grape must in Apulia, Southern Italy. Int J Microbiol 2014
Trivedi P, Leach JE, Tringe SG, Sa T, Singh BK (2020) Plant–microbiome interactions: from community assembly to plant health. Nat Rev Microbiol 18.11: 607–621
Upreti K, Murti G (2010) Response of grape rootstocks to salinity: changes in root growth, polyamines and abscisic acid. Biol Plant 54:730–734
Vadakattu GV, Bramley RG, Greenfield P, Yu J, Herderich M (2019) Vineyard soil microbiome composition related to rotundone concentration in Australian cool climate ‘peppery’Shiraz grapes. Front Microbiol 10:1607
Van Leeuwen C, Seguin G (2006) The concept of terroir in viticulture. J Wine Res 17:1–10
Van Zyl JL (1984) Interrelationships among soil water regime, irrigation and water stress in the grapevine (Vitis vinifera L.). Stellenbosch University, Stellenbosch
Volpe V, Chitarra W, Cascone P, Volpe MG, Bartolini P, Moneti G et al (2018) The association with two different arbuscular mycorrhizal fungi differently affects water stress tolerance in tomato. Front Plant Sci 9:1480
Wei Z, Gu Y, Friman V-P, Kowalchuk GA, Xu Y, Shen Q, Jousset A (2019) Initial soil microbiome composition and functioning predetermine future plant health. Sci Adv 5:eaaw0759
Whiteman S, Jaspers M, Stewart A, Ridgway H (2003) Identification of potential sources of Phaeomoniella chlamydospora in the grapevine propagation process. Phytopathol Mediterr 43:152
Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S et al (2015) The soil microbiome influences grapevine-associated microbiota. MBio 6:e02527–e02514
Zhang H, Xu N, Li X, Long J, Sui X, Wu Y et al (2018) Arbuscular mycorrhizal fungi (Glomus mosseae) Improves growth, photosynthesis and protects photosystem ii in leaves of Lolium perenne L. in cadmium contaminated soil. Front Plant Sci 9:1156
Zilber-Rosenberg I, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. Fems Microbiol Rev 32:723–735
Acknowledgements
The Fig. 6 was created with BioRender.com.
Funding
Part of the work was supported by Settesoli winery S.C.A. Menfi (AG) Sicily (IT) and by the BIOPROME in the frame of the DiBio project founded by the Italian Ministry of Agriculture.
Author information
Authors and Affiliations
Contributions
L.N. and W.C. designed the experimental system, carried out the wet lab experiments, analysed data and wrote the manuscript draft. D.T., L.N. and W.C. worked out vine roots system. D.T. analysed wines sensory description. D.T., A.G, L.M. and G.G. helped to design the experiments, contributed to the writing and carefully revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Additional information
Responsible Editor: Birgit Mitter.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
D. Tomasi and Walter Chitarra equally contributed as senior authors.
The SRA accession numbers of the NGS reported in this paper are deposited in NCBI under the BioProject PRJNA655455; BioSample SAMN15735114 and SAMN15735115; SRA accession SRR12436974 and SRR12436973.
Supplementary Information
Figure S1.
PCoA representing the four soil samples analysed for 16S sequences (n=3). (PPTX 2244 kb)
Figure S2.
NMDS algorithm based on Bray-Curtis distances matrixes were used to reduce into a bi-dimensional scaling data obtained for bacteria (A), fungi (B) and the overall microbial community (C) (n=3). (PPTX 315 kb)
Figure S3.
Cluster heatmap built using data of both 16S and ITS amplicon sequencing to highlight relationship among samples. (PDF 36 kb)
Figure S4.
Significant co-occurrence network of bacterial communities and interactions among OTUs in soil samples. The nodes are sized according to degree of connection (Supplementary Table 11). Green lines indicate that there are co-occurrence interactions and red lines indicate mutualistic exclusions (n=3). (PPTX 106 kb)
Figure S5.
Significant co-occurrence network of fungal communities and interactions among OTUs in soil samples. The nodes are sized according to degree of connection (Supplementary Table 12). Green lines indicate that there are co-occurrence interactions and red lines indicate mutualistic exclusions (n=3). (PNG 98902 kb)
Figure S6.
Significant co-occurrence network of both bacterial and fungal communities considered together and interactions among OTUs in soil samples. The nodes are sized according to degree of connection (Supplementary Table 13). Green lines indicate that there are co-occurrence interactions and red lines indicate mutualistic exclusions (n=3). (PNG 207531 kb)
Figure S7
(PNG 222690 kb)
ESM 1
(DOCX 268 kb)
ESM 2
(XLS 1292 kb)
Rights and permissions
About this article
Cite this article
Nerva, L., Moffa, L., Giudice, G. et al. Microscale analysis of soil characteristics and microbiomes reveals potential impacts on plants and fruit: vineyard as a model case study. Plant Soil 462, 525–541 (2021). https://doi.org/10.1007/s11104-021-04884-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-021-04884-2