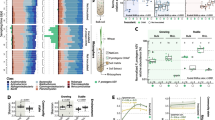
Abstract
Aims
Plant growth promoting bacteria (PGPB) have been used on crops for years, but inoculants that are efficient in some locations may not be efficient in others. Here, we applied classical invasion ecology theory to PGPB inoculation in order to identify patterns that can be used to predict plant growth promoting (PGP) efficiency. The hypotheses that the inoculant that causes most impact will be the most efficient PGPB, and that the most invasible locations would have higher PGP efficiency, were tested. We also aim to present our statistical approach to analyze SIMPER results.
Methods
Using next generation sequencing targeting the 16S rDNA gene in metagenomics samples, we analyzed samples of pre-planting bulk soil and rhizosphere of inoculated maize plants. Bacterial communities of inoculated plants were compared to the non-inoculated controls, in order to estimate the inoculant invasion impact. Crop yield was compared to different indexes, and a novel data exploration approach was employed.
Results
The most efficient inoculant was not the most invasive, and a nutrient per diversity ratio was unable to predict inoculant efficiency or invasion impact. However, the efficient inoculation treatment presented an enrichment of specific pre-planting taxa.
Conclusions
Invasion ecology frameworks could not anticipate field results of inoculated plants. Nonetheless, our data exploration approach, which is explained in detail, can be useful to raise new hypothesis and improve the visualization of dissimilarity data.






Similar content being viewed by others
References
Alves BJR, Boddey RM, Urquiaga S (2003) The success of BNF in soybean in Brazil. Plant Soil 252:1–9
Ambrosini A, de Souza R, Passaglia LMP (2016) Ecological role of bacterial inoculants and their potential impact on soil microbial diversity. Plant Soil 400:193–207. https://doi.org/10.1007/s11104-015-2727-7
Arruda L, Beneduzi A, Martins A et al (2013) Screening of rhizobacteria isolated from maize (Zea Mays L.) in Rio Grande do Sul state (South Brazil) and analysis of their potential to improve plant growth. Appl Soil Ecol 63:15–22. https://doi.org/10.1016/j.apsoil.2012.09.001
Bashan Y, De-Bashan LE, Prabhu SR, Hernandez J-P (2014) Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant Soil 378:1–33. https://doi.org/10.1007/s11104-013-1956-x
Berg G, Zachow C, Müller H et al (2013) Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy 3:648–656. https://doi.org/10.3390/agronomy3040648
Caporaso JG, Lauber CL, Walters WA et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108:4516–4522. https://doi.org/10.1073/pnas.1000080107/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1000080107
Castro-Sowinski S, Herschkovitz Y, Okon Y, Jurkevitch E (2007) Effects of inoculation with plant growth-promoting rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiol Lett 276:1–11. https://doi.org/10.1111/j.1574-6968.2007.00878.x
Chowdhury SP, Dietel K, Rändler M et al (2013) Effects of bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS One 8:e68818. https://doi.org/10.1371/journal.pone.0068818
Clarke KR (1993) Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143
Clarke KR, Warwick RM (2001) Change in marine communities: an approach to statistical analysis and interpretation, 2nd editio. PRIMER-E, Plymouth
Compant S, Clément C, Sessitsch A (2010) Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem 42:669–678. https://doi.org/10.1016/j.soilbio.2009.11.024
Davis MA, Pelsor M (2001) Experimental support for a resource-based mechanistic model of invasibility. Ecol Lett 4:421–428. https://doi.org/10.1046/j.1461-0248.2001.00246.x
Davis MA, Grime JP, Thompson K (2000) Fluctuating resources in plant communities: a general theory of invasibility. J Ecol 88:528–534
Forrest BM, Taylor MD (2002) Assessing invasion impact: survey design considerations and implications for management of an invasive marine plant. Biol Invasions 4:375–386. https://doi.org/10.1023/A:1023613428351
Freedman ZB, Romanowicz KJ, Upchurch RA, Zak DR (2015) Differential responses of total and active soil microbial communities to long-term experimental N deposition. Soil Biol Biochem 90:275–282. https://doi.org/10.1016/j.soilbio.2015.08.014
Gilbert GS, Clayton MK, Handlesman J, Parke J (1996) Use of cluster and discriminant analysis to compare rhizosphere bacterial communities following biological perturbation. Microb Ecol 32:123–147
Gitipour A, El Badawy A, Arambewela M et al (2013) The impact of silver nanoparticles on the composting of municipal solid waste. Environ Sci Technol 47:14385–14393
Haegeman B, Moriarty J, Neal P et al (2013) Robust estimation of microbial diversity in theory and in practice. ISME J 7:1092–1101. https://doi.org/10.1038/ismej.2013.10
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4(1):9
Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH (2012) Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. https://doi.org/10.1038/nrmicro2795
Hierro JL, Maron JL, Callaway RM (2005) A biogeographical approach to plant invasions: the importance of studying exotics in their introduced and native range. J Ecol 93:5–15. https://doi.org/10.1111/j.1365-2745.2004.00953.x
Hungria M, Campo RJ, Souza EM, Pedrosa FO (2010) Inoculation with selected strains of Azospirillum brasilense and A. Lipoferum improves yields of maize and wheat in Brazil. Plant Soil 331:413–425. https://doi.org/10.1007/s11104-009-0262-0
Kassen R, Buckling A, Bell G, Rainey P (2000) Diversity peaks at intermediate productivity in a laboratory microcosm. Nature 406:508–512
Linting M, Meulman JJ, Groenen PJF, Van der Kooij AJ (2007) Nonlinear principal components analysis: introduction and application. Psychol Methods 12:336–358. https://doi.org/10.1037/1082-989X.12.3.336
Mallon CA, Elsas JD Van, Sales JF (2015) Microbial invasions : the process, patterns, and mechanisms. Trends Microbiol 23:719–729
Margherita G, Osborne B (2009) Assessing the impact of plant invasions on soil seed bank communities : use of univariate and multivariate statistical approaches. J Veg Sci 20:547–556. https://doi.org/10.1111/j.1654-1103.2009.01054.x
Mills DK, Entry JA, Voss JD, et al (2006) An assessment of the hypervariable domains of the16S rRNA genes for their value in determining microbial community diversity : the paradox of traditional ecological indices. 57:496–503. doi:https://doi.org/10.1111/j.1574-6941.2006.00135.x
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a web browser. BMC Bioinf 12:385. https://doi.org/10.1186/1471-2105-12-385
Otieno BN, Culhane J, Germaine KJ et al (2013) Screening of large collections of plant associated bacteria for effective plant growth promotion and colonisation. Asp Appl Biol 120:23–28
Owen D, Williams AP, Griffith GW, Withers PJA (2015) Use of commercial bio-inoculants to increase agricultural production through improved phosphrous acquisition. Appl Soil Ecol 86:41–54. https://doi.org/10.1016/j.apsoil.2014.09.012
Parker IM, Simberloff D, Lonsdale WM et al (1999) Impact : toward a framework for understanding the ecological effects of invaders. Biol Invasions 1:3–19
Purcell SW, Clarke KR, Rushworth K, Dalton SJ (2014) Defining critical habitats of threatened and endemic reef fishes with a multivariate approach. Conserv Biol 28:1688–1698. https://doi.org/10.1111/cobi.12343
Ramette A (2007) Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160. https://doi.org/10.1111/j.1574-6941.2007.00375.x
Rees GN, Baldwin DS, Watson GO, Perryman S (2004) Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek 86:339–347
Ryan RP, Germaine K, Franks A et al (2008) Bacterial endophytes: recent developments and applications. FEMS Microbiol Lett 278:1–9. https://doi.org/10.1111/j.1574-6968.2007.00918.x
Satomi M (2014) The family Shewanellaceae. In: Rosenberg E, Delong EF, Thompson F (eds) The prokaryotes, 4th edn. Springer, Berlin, pp 597–619
Sparks DL, Page AL, Helmke PA, Loeppert RH (1996) Methods of soil analysis: part 3. Chemical methods and processes. Soil Science Society of America, Madison, USA
Stevens JL, Olson JB (2013) Invasive lionfish harbor a different external bacterial community than native Bahamian fishes. Coral Reefs 32:1113–1121. https://doi.org/10.1007/s00338-013-1072-7
Thomas T, Gilbert J, Meyer F (2012) Metagenomics - a guide from sampling to data analysis. Microb Inf Exp 2:3. https://doi.org/10.1186/2042-5783-2-3
van Elsas JD, Chiurazzi M, Mallon CA et al (2012) Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci U S A 109:1159–1164. https://doi.org/10.1073/pnas.1109326109
Wang J, Linnenbrink M, Künzel S, et al (2014) Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. PNAS 111:1–8. doi:https://doi.org/10.1073/pnas.1402342111
Wilkins D, Lauro FM, Williams TJ et al (2013) Biogeographic partitioning of Southern Ocean microorganisms revealed by metagenomics. Environ Microbiol 15:1318–1333. https://doi.org/10.1111/1462-2920.12035
Zelezniak A, Andrejev S, Ponomarova O et al (2015) Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci U S A 112:E7156. https://doi.org/10.1073/pnas.1522642113
Acknowledgements
This work was financed by grant and fellowships from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/Brazil) and Instituto Nacional de Ciência e Tecnologia (INCT) da Fixação Biológica do Nitrogênio (Brazil) for Brazilian researchers; Deutscher Akademischer Austauschdienst (DAAD/Germany) for PBdC; Bielefeld Young Researchers Fund for SBC. The bioinformatics support of the BMBF-funded project “Bielefeld-Giessen Center for Microbial Bioinformatics – BiGi (Grant number031A533)” within the German Network for Bioinformatics Infra-structure (de.NBI) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Responsible Editor: Timothy Cavagnaro.
Electronic supplementary material
Fig. S1
Relative Phyla (p_) distribution per sample. Phyla legend is in the order of the most common to the rarest. L = Londrina; P = Ponta Grossa; M = Marechal Cândido Rondon; 0 = pre-planting; 1 = non-inoculated control; 2 = Azospirilum brasilisense Ab-V5; 3 = Achromobacter sp. VC36; 4 = Pseudomonas sp. 4311; 5 = Pseudomonas sp. 4312; a = first replicate; b = second replicate (GIF 49 kb)
Fig. S2
SIMPER percentages from the treatment-control pairs, at each taxonomic level. Color gradient shows lower values in red, higher values in green. L = Londrina; P = Ponta Grossa; M = Marechal Cândido Rondon; 2 = Azospirillum brasilense Ab-V5; 3 = Achromobacter sp. VC36; 4 = Pseudomonas sp. 4311; and 5 = Pseudomonas sp. 4312 (GIF 38 kb)
Fig. S3
SIMPER dissimilarity data from Online Resource 4 treated under a NMDS instead of a PCA. (a) NMDS based on Euclidian distance, with an output similar to the SIMPER-PCA approach (b) Shepard plot showing fit and stress level for the NMDS solution based on Euclidean distance (c) NMDS based on the Bray-Curtis dissimilarity, calculating a dissimilarity metric on dissimilarity data, showing clear artifacts (d) Shepard plot showing fit and stress level for the NMDS solution based on Bray-Curtis dissimilarity. Red = Londrina (L); Blue = Ponta Grossa (P); black = Marechal Cândido Rondon (M); full circle = Azospirillum brasilense Ab-V5 (2); empty circle = Achromobacter sp. VC36 (3); empty square = Pseudomonas sp. 4311 (4); and full square = Pseudomonas sp. 4312 (5) (GIF 25 kb)
Fig. S4
Example of the SIMPER-PCA approach at OTU level without transforming the signal as suggested in the text. Different numbers represent different OTUs. (GIF 34 kb)
Fig. S5
Histograms for frequencies of the average dissimilarities calculated from SIMPER tests as given by the PAST output (a) and with the signal modification (b). These values are presented on Online Resource 4. The curved line shows the theoretical normal distribution for the dataset. The lower value for the central bin in part (b), that might resemble a dent in the normal distribution, shows that there are not many taxa that explain very little variance. This happens because of the minimum frequency OTU cut off. OTUs with less than 200 occurrences were removed from the dataset, exactly because they would explain little variance and increase background noise. Without this cut-off, the central bin in this figure would be much higher, total variance explained would be lower, but the clustering of the datapoints in the SIMPER-PCA approach would not differ drastically. (GIF 11 kb)
Online Resource 1
Full dataset of crop productivity. At location column: L = Londrina; P = Ponta Grossa; M = Marechal Cândido Rondon. At inoculant column: 1 = non-inoculated control; 2 = Azospirillum brasilense Ab-V5; 3 = Achromobacter sp. VC36; 4 = Pseudomonas sp. 4311; and 5 = Pseudomonas sp. 4312. At block column: blocks one to four, for ANOVA analysis. (XLSX 15 kb)
Online Resource 2
Full sequencing results (absolute values) for every read at all at taxa levels. At sample column: L = Londrina; P = Ponta Grossa; M = Marechal Cândido Rondon, 1 = non-inoculated control; 2 = Azospirillum brasilense Ab-V5; 3 = Achromobacter sp. VC36; 4 = Pseudomonas sp. 4311; and 5 = Pseudomonas sp. 4312; a = first replicate; b = second replicate. (XLSX 869 kb)
Online Resource 3
Some of the SIMPER percentages from the treatment-control pairs, at OTU level. The full signaled results, used for Fig. 4, are presented on Online Resource 4. Av. dissim. = taxa average dissimilarity; contrib. % = contribution of that taxa to total dissimilarity; Cumul. % = cumulative total dissimilarity; Mean abundances = normalized and transformed counts of each taxa for control and treatments; p_ = Phyla; c_ = Class; o_ = Order; f_ = Family; g_ = genera. (XLSX 17 kb)
Online Resource 4
Signal transformed SIMPER dissimilarities from the treatment-control pairs, at OTU level, used to create Fig. 4 and Fig. S3. L = Londrina; P = Ponta Grossa; M = Marechal Cândido Rondon. At inoculant column: 1 = non-inoculated control; 2 = Azospirillum brasilense Ab-V5; 3 = Achromobacter sp. VC36; 4 = Pseudomonas sp. 4311; and 5 = Pseudomonas sp. 4312.; p_ = Phyla; c_ = Class; o_ = Order; f_ = Family; g_ = genera. (XLSX 360 kb)
Online Resource 5
SIMPER values for each taxon at pre-planting conditions, using Londrina as the standard compared to Marechal Cândido Rondon (a) and Ponta Grossa (b), both at OTU level. (XLSX 339 kb)
ESM 1
(PDF 126 kb)
Rights and permissions
About this article
Cite this article
da Costa, P.B., de Campos, S.B., Albersmeier, A. et al. Invasion ecology applied to inoculation of plant growth promoting bacteria through a novel SIMPER-PCA approach. Plant Soil 422, 467–478 (2018). https://doi.org/10.1007/s11104-017-3492-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3492-6