Abstract
Background
Plant-soil interaction is central to human food production and ecosystem function. Thus, it is essential to not only understand, but also to develop predictive mathematical models which can be used to assess how climate and soil management practices will affect these interactions.
Scope
In this paper we review the current developments in structural and chemical imaging of rhizosphere processes within the context of multiscale mathematical image based modeling. We outline areas that need more research and areas which would benefit from more detailed understanding.
Conclusions
We conclude that the combination of structural and chemical imaging with modeling is an incredibly powerful tool which is fundamental for understanding how plant roots interact with soil. We emphasize the need for more researchers to be attracted to this area that is so fertile for future discoveries. Finally, model building must go hand in hand with experiments. In particular, there is a real need to integrate rhizosphere structural and chemical imaging with modeling for better understanding of the rhizosphere processes leading to models which explicitly account for pore scale processes.
Similar content being viewed by others
Introduction
“We know more about the motion of celestial bodies than about the ground underneath our feet” (Leonardo da Vinci). Although this statement is approximately 500 years old, it is still valid for the soil close to the root, the rhizosphere. In the rhizosphere plant roots interact with the soil, altering its physical, chemical and biological properties (Hinsinger et al. 2009). This process has been shown to affect the ability of plant roots to extract water and nutrients from the soil, in particular when such resources are scarce (Hinsinger et al. 2009).
Root-soil interactions also affect the pore structure within the rhizosphere in a complex way, which is still poorly understood and may depend on a variety of different factors. Existing studies suggest an increase in soil density around the roots (Aravena et al. 2014; Bruand et al. 1996; Dexter 1987b). However, soil densification around the roots may not be the general rule. For instance Feeney et al. (2006) showed that plant roots and associated microorganisms increase soil porosity. Whalley et al. (2005) measured a greater number of large pores in aggregates collected from the rhizosphere. On the other hand, Daly et al. (2015a) found lower macroporosity in planted samples compared to unplanted ones. Additionally it has to be kept in mind that as transpiration increases and the soil dries, roots shrink and may partially lose contact with the soil (Huck et al. 1970), creating large air-filled pores around the roots (Carminati et al. 2013).
The mechanisms controlling the temporal dynamics of structural changes in the rhizosphere are poorly understood. Even less is known about how rhizosphere structure affects water and nutrient fluxes into the roots. So far, the mathematical description of root-soil interactions has been impeded by our inability to study such interactions in situ, i.e., in undisturbed soil environments (Hutchings and John 2004; Pierret et al. 2007). However, we now have a set of existing and emerging tools and techniques that enable us to do this. Thus, in this review we will discuss the development of mathematical models that explicitly take into consideration the structure of the pore space around the roots and how it is affected by root growth, exudates, root hairs and soil shrinking-swelling cycles. We will also discuss emerging experimental techniques that are necessary to make these models rigorous, experimentally validated and scientifically useful. We will highlight current achievements and major challenges in understanding the relation between rhizosphere structure and its function in controlling water and nutrient uptake.
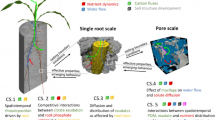
Specifically, we focus on imaging root-soil interactions with a drive towards producing image based, fully calibrated, predictive models which integrate processes from micro-meter to decimeter-scales, and across temporal scales from seconds to months. We will aim our discussion at situations where plants are grown in the soil in pots. This is the scale at which most studies are carried out and offers the most potential for future progress using modelling to integrate data generated by new imaging tools. To make this progress, several challenges need to be overcome. These relate to improvements in image quality and resolution, as well as integration of physical, chemical and biological techniques to fully understand processes in the rhizosphere. Technological advances alone are not sufficient. Real advances in our understanding will only be achieved if these data can be integrated, correlated, and used to parameterize and validate image based and mechanistic models. Clearly every model, image based or not, has a set of assumptions in it and no model is ever perfect, fully mechanistic and fit to answer every question in the particular area. Rather, mathematical modeling at its best serves to guide future experimental investigations to increase the predictive power of the models.
We will concentrate on current advances in rhizosphere imaging and how these can aid the development of models, and we highlight the need to further integrate imaging and modeling approaches in this area. In particular we will point out where we think major knowledge gaps in imaging and modeling integration lie. Specifically we review pore- to root-scale effects in two areas: (i) imaging root-induced physical/structural and chemical processes in the rhizosphere and (ii) image-based modeling. Our specific focus is water movement and the transport of strongly-bound heavier nutrients, such as phosphate (P), and their interaction with root structures and the overall root system architecture. In this context we will discuss challenges that we face in upscaling rhizosphere processes. Processes on a scale smaller than a single root, and processes at the field scale are not considered as these have been comprehensively reviewed elsewhere, for example Peret et al. (2009) and Vereecken et al. (2015), respectively. The integration of knowledge and identification of knowledge gaps for mathematical modeling is the focus of our review.
Existing work on rhizosphere imaging and modeling
There are some excellent recent reviews that deal with issues related to our paper. However, they all deal with plant-scale structural imaging of processes, i.e., they do not address the challenges associated with pore-scale imaging, multi-scale imaging, and correlative chemical mapping of the rhizosphere and associated modeling techniques. The use of X-ray computed tomography methods to probe root-soil macroscopic interactions has recently been reviewed by Helliwell et al. (2013); Jones et al. (2013); Mooney et al. (2012). We will build on these reviews and focus on rhizosphere-specific aspects, i.e., high resolution imaging of root and soil architecture and interactions within the changing rhizosphere environment with specific relevance to mathematical modelling. The review of Downie et al. (2014) covers the challenges and opportunities facing researchers and practitioners interested in fast phenotyping of root systems. Their review discusses the potential of various techniques, including the use of transparent soil, to provide better understanding of root-soil interactions. Other reviews in this field deal with modelling of rhizosphere and plant-soil interactions (Hinsinger et al. 2011; Dunbabin et al. 2013), mycorrhizae (Treseder 2013), mycorrhizal nitrogen uptake (Hodge and Storer 2015), and transport processes in porous media (Wildenschild and Sheppard 2013). Finally, there is a collection of articles published in edited books (Anderson and Hopmans 2013; Bengough 2012; Timlin and Ahuja 2013) covering issues related to this review, such as neutron and X-ray imaging.
The key scientific challenges identified in all of the reviews above are mainly focused on our ability to observe root architectural morphology, soil structure and chemical composition over limited spatial and temporal scales, often with techniques targeting a single property or process. The translation of this known organization of system information across scales is thus the challenge that needs to be met by mathematical and computational methodologies and their development.
Modelling rhizosphere processes: state of the art
Rhizosphere modelling has a long-standing history dating back to the 1960s (Barber 1984; Olsen and Kemper 1968; Tinker and Nye 2000). Most of this research has concentrated on modelling rhizosphere chemical changes and water dynamics and has largely focused on individual elements. At best two aspects (two elements of water-nutrient interaction) have been integrated, but this has not always been the case. For example, De Willigen and Van Noordwijk (1984); Van Noordwijk and De Willigen (1984) presented mathematical formulations and steady-state solutions for diffusive transport of oxygen inside roots in relation to root-soil contact, in which oxygen diffusion into roots was limited either by soil particles or water films. They showed that root-soil contact considerably affects the partial oxygen pressure required for aerobic respiration, which was higher for soil grown roots than for those in well-stirred nutrient solutions. In a series of other papers they derived simple approximations of analytical solutions for a “zero-sink”Footnote 1 uptake of nutrients by a plant root with transport by diffusion and mass flow (De Willigen and Van Noordwijk 1994a, b). They then qualitatively compared this theoretical understanding with experiments where root-soil contact was altered by varying soil bulk density. Their work set an early theoretical framework for how physical processes around roots can be considered in root system and crop growth models. However, the underlying theory at the time was essentially centred on simplified assumptions of the physical conditions in the rhizosphere and did not capture the heterogeneity we are now able to observe. A good overview of these early endeavours is presented by Fitter (2002).
The new state of the art approach to modelling root-soil interaction is based on root system architecture, i.e., models which take into account the specifics of root system architecture at the expense of high computational cost (Dunbabin et al. 2013; Ge et al. 2000; Pages 2011). While root system architecture has in the past been derived from a range of computational models, it is now possible to measure it in situ (i.e., in the soil) using imaging techniques such as magnetic resonance imaging (MRI), X-ray computed tomography (X-ray CT) and neutron tomography, see Carminati et al. (2010); Gregory et al. (2003); Koebernick et al. (2014); Metzner et al. (2015); Mooney et al. (2012); Moradi et al. (2011); Oswald et al. (2008) as a good starting point for the literature. These images can be utilised to build an image-based model for water and/or nutrient uptake by the root system. However, surprisingly few models utilising this imaging information exist. The root system is usually represented in the nutrient mass balance equation as a synthetic architecture or image-derived sink term, i.e., the specific root architectural information is averaged over a given soil volume to build a sink term (Dunbabin et al. 2013). This is the case for all/most models, such as R-SWMS, discussed by Dunbabin et al. (2013). Obviously the need for this averaging arises primarily from the lack of computational resources available to most rhizosphere modelling groups. In particular the memory requirements for image based modelling can easily exceed 100Gb of RAM and, in order to ensure that models can be run over a couple of days, multi-node supercomputing resources are essential. It is undoubtedly clear that these architectural averaged models, such as R-SWMS, greatly help to test our understanding of system function and, due to their relatively low computational cost, are easy to access and run on standard computational platforms (PCs) (Koebernick et al. 2015). However, it is important to be aware that there are limitations to their use and there are some serious assumptions inherent in the models that might limit their applicability. For example, it is almost impossible to include pore-scale rhizosphere morphological effects in these models with accuracy. We are not aware of any effort in the past to do this, except Heppell et al. (2015) who did include the root hair morphological effect in a soil profile scale model in a simple parametric manner.
The architectural modelling approach now includes direct time dependent and 3D-space explicit computations of plant P uptake (Leitner et al. 2010b). In these models root surfaces are explicitly represented without any a priori averaging, and boundary conditions are applied for the root-soil interface domain rather than the root system being represented by a volume-averaged sink term. Following this development, Keyes et al. (2013) imaged and conducted image-based pore-scale modelling of plant P uptake by root hairs in which the root, root hair, and soil particle surfaces were all explicitly accounted for, resulting in the first ever image based rhizosphere model that included such structural information. A hierarchy of different models is ultimately necessary since high levels of detail and complexity in models require computational resources and time, which contraindicate high-throughput approaches. Thus, detailed explicit models are perhaps best utilised to verify, test and validate faster and less complex models that include significant simplifications and approximations. Thus, the high-detail ‘gold standard’ models help to make sure that simplifications do not introduce mathematical artefacts and distort scientific interpretation. However, the emergence of these models also highlights the need for more accurate and detailed characterisation of the soil chemistry; for example, buffer-power-style equilibrium characterisation of bulk soil chemistry is not very informative for the pore scale processes as described in Keyes et al. (2013).
The image based modelling of soil hydrological and petrological processes, and in particular pore scale modelling where specific aggregate structure is accounted for, has a somewhat longer track record than imaging and modelling of the rhizosphere (Joekar-Niasar et al. 2012; Wildenschild and Sheppard 2013). Various authors give excellent overviews of all the X-ray based CT and XRF techniques and modelling that have been applied to study porous media (such as soil), with an emphasis on hydrological and petrological studies (Blunt 2001a; Blunt et al. 2013; Lombi and Susini 2009; Wildenschild and Sheppard 2013). One particularly major challenge is the identification of all four rhizosphere phases in image data, i.e., air, water, soil minerals, organic matter (roots, mucilage and soil organic matter), and this has undoubtedly impeded the development of mathematical models. The key scientific unknown is how these phases interact in the soil pore space and how they quantitatively and qualitatively influence soil processes such as plant nutrient and water uptake, mineralisation/mobilisation of nutrients, and feedback processes including release of substances, growth and tissue differentiation. A model of root water uptake including mucilage dynamics in the rhizosphere has been recently introduced in a series of articles by Carminati (2012); Carminati et al. (2010); Ghezzehei and Albalasmeh (2015); Kroener et al. (2014). In these modelling studies, the rhizosphere hydraulic properties differ from those of the bulk soil and vary over time during drying and wetting cycles. These models were derived based on time-series neutron radiographs of plants grown in sandy soil with low soil organic matter. The imaging revealed the water content in the rhizosphere and in the adjacent bulk soil. The models showed how small-scale processes across the rhizosphere impact on root water uptake and the relations between bulk soil water potential, root water potential and transpiration rates. In future, these models should be implemented in a three-dimensional setting for a range of soil types and textures with different soil organic matter content and water saturation, taking into account the root structure and architecture. This is clearly the future challenge since not only does one need to understand processes at the soil pore scale, but these results must be translated across scales accurately and reliably in order to synthesise new scientific knowledge.
Brief review of structural imaging
In this review we use the term structural imaging in reference to methods which directly visualise and quantify structure and morphology associated with plant-soil interactions. The current techniques available, such as X-ray CT, neutron CT and MRI, allow for a step change in our understanding by enabling explicit spatial characterisation of the dynamics of soil structure in the vicinity of the roots (Carminati et al. 2013; Pagenkemper et al. 2013; Singh and Grafe 2010), see Fig. 1, as well as detailed characterisation of root architecture (Koebernick et al. 2014).
Left: Surface area of a single root induced biopore including connected lateral “secondary”branching channels in the rhizosphere. Left: Single root with branching secondary laterals. Middle: Pore skeleton (medial axis) of biopore network with colors indicating local channel width (burn number) from red indicating very narrow channel diameters over yellow, green, blue to purple colors corresponding to increasingly wider channels. Right: in situ sample of a single root with branching secondary laterals
Structural imaging techniques enable us to visualise microscopic heterogeneities of soil in the proximity of the root surface (such as structural changes or changes in water content) and how they evolve over time, in addition to observing macroscopic changes in the root structure in natural soils (Mooney et al. 2012). For example, Grose et al. (1996) applied clinical X-ray CT to quantify the heterogeneity in water content around wheat roots and used these data to identify regions around root systems that were more or less favourable for soil-borne fungal pathogens such as Gaeumanomyces graminis var. tritici and Rhizoctonia solani. More recently, and with the development of X-ray CT systems capable of obtaining data at higher spatial resolution, data can now be obtained which quantify the changes in soil structure around roots. For example, Aravena et al. (2010); Aravena et al. (2011) used synchrotron data to show soil (clay loam aggregates) compaction around roots (sweet pea and sunflower), demonstrating that inter-aggregate porosity decreased within 300 μm from the root surface. This compaction resulted in an increase in contact between aggregates and numerical modelling was employed to show that the unsaturated hydraulic conductivity around roots might increase as a result. This means that counter intuitively, water flow may be locally enhanced due to root-induced compaction of aggregated soil. Root-soil contact is another important characteristic, which influences water and nutrient uptake (De Willigen and Van Noordwijk 1994a; b; Nye 1994). X-ray CT offers the opportunity to quantify root-soil contact and to identify how it evolves over time as the root grows and is affected by soil properties. An example of this was presented by Schmidt et al. (2012) who developed a method to quantify root-soil contact from X-ray CT data. Quantifying contact areas is particularly challenging in X-ray CT due to partial volume effects,Footnote 2 yet using phantoms of known geometry and dimensions to calibrate the imaging of contact between two bodies they showed that quantification with ~97 % accuracy can be achieved. They demonstrated that for young seedlings, minor differences in the macro-porosity of the bulk soil can have substantial effect on root-soil contact.
In addition to the effect of near-root soil compression on hydraulic functions, it is important to probe mechanical aspects of root-soil interaction. For example, as a consequence of deformation, the mechanical properties of the rhizosphere soil will change. This will impact upon the penetration of secondary lateral roots into the rhizosphere of the primary root. It is also well known that root proliferation and root architecture are controlled by soil mechanical strength and its spatial heterogeneity resulting in the situation that roots are seldom presented with a homogeneous mechanical environment (Groenevelt et al. 1984). There are various ways in which roots respond to high mechanical impedance, e.g., by sloughing of border cells and the release of exudates. Root thickening is another strategy used to penetrate dry and hard soil, resulting in reduction of stress in front of the root apex and lower resistance to root elongation (Bengough et al. 2006).
To model such processes (e.g., using finite element approaches), it is crucial to determine the mechanical properties of the soil and their changes with root growth in situ at the individual root scale. Image correlation techniques such as Digital Image Correlation (DIC) and Digital Volume Correlation (DVC) are promising tools to investigate the mechanics of root-soil interaction. DIC was developed in the 1980s alongside the advent of digital image processing and affordable numerical computing (Peters and Ranson 1982). In DIC the deformation on planar sample surfaces (i.e., of a tensile test coupon) is quantified by tracking an inherent or user-applied pattern between sequential digital images acquired during loading. With determination of suitable parameters and consideration of the various sources of error, it is thus possible to derive full-field strain data without resorting to invasive and/or sparse methods such as strain gauging (Bay 2008). The implementation of DIC is very similar to that of the particle image velocimetry (PIV) approaches used in experimental fluid mechanics (Willert and Gharib 1991). Briefly, comparison is made between reference and deformed sample states by subdividing images of the respective surface pattern into sub-regions. For each sub-region, the affine transform is determined that maps each sub-region between reference and deformed positions. Various schemes are available to achieve this, although the standard approach is to minimise an objective function to determine the degree of similarity in pattern between the reference sub-region and each test location in the deformed image (Pan et al. 2009). Once the displacement vector of each sub-region is known, it is possible to estimate the strain at any point by computing the gradient of the displacement field. For the interested reader, a full review of DIC methods is provided in Pan et al. (2009). The widespread adoption of industrial micro X-ray CT imaging has led to the extension of DIC to the 3D case, known as digital volume correlation (DVC). Bay et al. (1999) were the first to extend the DIC approach to 3D data acquired using a bench-top CT scanner, applying the technique to samples of trabecular bone in simple uniaxial compression. Since this first demonstration, in which sub-voxel precision in displacement measurement was achieved, variations of the method have been applied to study a diverse range of materials including sand (Hall et al. 2010), woods (Forsberg et al. 2008), sugar (Forsberg and Siviour 2009), metals (Morgeneyer et al. 2013), gels (Huang et al. 2011), rock (Lenoir et al. 2007), engineering composites (Brault et al. 2013), and foams (Roux et al. 2008). Because DVC and DIC provide full-field deformation information and are physically non-invasive, they are highly promising techniques for investigating the mechanics of soil and root systems whose opacity, heterogeneity and complexity make other strain measurement approaches unfeasible. Bengough et al. (2010) have used PIV to study the root growth and rhizosphere displacement in ballotini/agar using confocal laser scanning microscopy images. Vollsnes et al. (2010) used PIV and optical images of rhizoboxes to measure soil displacement around maize roots, finding significant differences in deformation field between wild type and a root with root cap removed resulting in lower levels of mucilage in the rhizosphere. DVC has recently been applied to X-ray CT data of soil core samples, allowing the mapping of strain localisation related to hydrologically-driven shrinking, swelling and uniaxial compression, revealing very complex and heterogeneous deformation patterns (Peth et al. 2010). By iteratively mapping the reference tomogram (non-deformed state) to the tomogram of the deformed state, the authors were able to derive the Lagrangian strain tensor, which is a complete representation of the state of strain at a point (including volumetric, and shear components). Such data can be used to define stress–strain-relationships and thus parameterize mechanical models simulating root penetration.
In addition to mechanical deformation, microorganisms in the rhizosphere contribute to changes in the structure. Recent work has demonstrated that microbial growth can have a direct impact on the structural development of soils (Helliwell et al. 2014a; Nunan et al. 2006a). Helliwell et al. (2014b); Nunan et al. (2006b) demonstrated that within weeks of inoculation, pore volumes of individual pores as well as the bulk porosity of aggregates could be significantly increased by microbial activity. However, this came at the expense of using large amounts of glucose to promote the microbial activity. Specific bacterial habitats in terms of availability of decomposable substrate, oxygen and water will control mineralisation and mobilisation of nutrients and thus nutrient uptake by plant roots. To the best of our knowledge, little information exists with respect to the 3D spatial location of microbes within the rhizosphere. However, a method using transparent soil has recently been suggested for rhizosphere research that permits the use of normal light transmission microscopy (Downie et al. 2012, 2014). This method uses particles of a polymer (Nafion) which become ‘invisible’ following the addition of a solution with a matching refractive index. Particle sizes can be manipulated to obtain different structures for root growth in a 3D porous medium. This method can provide roots whose growth traits are comparable with soil-grown controls (Downie et al. 2012), but has the great advantage that light microscopy can be applied for visualisation of microbial colonization and distribution in the rhizosphere (Downie et al. 2014). Using multiple fluorescent signals in situ it is possible to study the growth and interactions of biological organisms in a physically complex soil-like environment. A clear disadvantage of transparent soil is that it is not soil, i.e., it does not have the chemical properties of a natural soil, even if the physical properties are closer to soil than to agarose gel. With the rhizosphere being a hotspot of microbial activity, the impact of rhizosphere microorganisms on soil structure development warrants further investigation.
The issue of imaging soil organic matter lies at the crossroads between structural and chemical imaging; the latter is the subject of the next section. However, we will discuss this issue here since, in terms of imaging technique, it is closer to structural imaging than chemical mapping. Ultimately, it is the synergy between these different techniques that will enhance our scientific understanding. The heterogeneous distribution of soil organic matter in the rhizosphere, and its potential as an energy source for microbes is largely unknown. Such information could significantly improve the simulation of microbial decomposition of soil organic matter. This is crucial for models, which are based on a 3D description of the pore space geometry, like the one recently developed by Monga et al. (2008). This was subsequently compared with experimental data to predict organic matter degradation in structured soil (Monga et al. 2014). In the absence of a method to directly visualise the spatial distribution of organic matter in soil, a particular strength of these modelling approaches is that they allow for scenario testing. For example, the effect of hypothetical distributions of organic matter in soil (i.e., size distributions of particulate SOM) on microbial activity can be assessed in order to formulate new hypotheses and insights for further experiments (Falconer et al. 2015). Visualising soil organic matter non-invasively, for example by X-ray CT, is difficult due to the low contrast between organic matter and other soil constituents, and the influence of partial volume effects if SOM is not clustered in sufficient quantity. Peth et al. (2014) used osmium tetroxide, which reacts with unsaturated C-bonds of organic compounds, to locate SOM in soil aggregates by absorption edge scanning at a synchrotron facility. Kravchenko et al. (2014) used preliminary particulate organic matter (POM) identification based upon grey-scale values, shape and sizes of POM pieces and conducted a discriminating analysis using statistical and geostatistical characterisation. They demonstrated that accurate quantification of POM inside aggregates could be achieved this way. Further development, such as staining methods that could make microorganisms and organic matter visible by non-invasive techniques, may bring us a step towards deriving spatially explicit input data for pore-scale modelling approaches.
Brief review of chemical mapping
In contrast to modeling and imaging, rhizosphere chemistry is a relatively well-studied area (Hinsinger et al. 2005, 2009; McNear 2013). In addition to simple field studies that compare chemical concentrations between bulk soil and rhizosphere soil, compartment-system approaches with a known position of the root-soil interface have contributed tremendously to process understanding (Neumann et al. 2009). The latter approach has been used based either on destructive sampling and application of conventional soil analysis using different extractants, or by using radio-labelled nutrients or stable isotopes. Temporal dynamics have been addressed in such systems by the installation of sensors at known distances from a root mat (Vetterlein and Jahn 2004). This approach can be adapted to use micro suction cups, optodes, redox electrodes or any other sensor with a sufficiently small form factor. Compartment systems provide only a linear geometry instead of the true radial geometry around a root and do not allow processes to be resolved along a developing root from the tip to more basal parts. Such resolution is possible using a rhizobox or root windows, wherein roots grow in the soil along a transparent plate which is either perforated to allow installation of sensors at certain positions along a root (Neumann et al. 2009), or which can be removed to allow direct contact of the visible root-soil interface with an imaging device (Dinkelaker et al. 1993). These chemical mapping systems always require a soil matrix whose texture enables good contact between the imaging device and the root-soil interface. This is important, because whether the device is a gel (agar, agarose, polyacrylamide), glass-fiber or paper filter, membrane, ion-exchange resin or foil, the species from the root-soil interface are brought into contact with the imaging device via diffusion (Neumann et al. 2009). A key advantage of these techniques is that one can image whole root systems, or at least large parts of root systems, in 2D. Temporal information can be obtained by repeating the procedure over a series of time points. The major drawback is that the gradients measured depend on the diffusive conditions, not only within the soil, but also within the device mediating the contact. Hence it is not only the sensor material properties which have an impact, but also soil moisture and sample exposure time. In addition, there is some uncertainty regarding the extent to which the conditions at the interface with a transparent plate are representative of roots that are entirely surrounded by soil. Roots that are present within the rhizobox, but not visible along the front plate, may further confound the results.
2D-imaging in rhizoboxes has been applied to study release of carbon or specific organic compounds by 14C application and/or chemical analyses of spatial resolved collected exudates with HPLC, HPLC-MS, capillary electrophoreses or GC-MS (Dessureault-Rompré et al. 2006; Haase et al. 2007; Neumann et al. 2009). Enzyme activity (acid phosphatase) was imaged based on dye-impregnated filter paper as early as 1992 by Dinkelaker and Marschner (1992). Recently this method has been rediscovered and extended to alkaline phosphatase under the term zymography (Spohn and Kuzyakov 2013). FeII-oxidation, FeIII-reduction, Mn-reduction, and Al-complexation have been studied using different dyes (Neumann et al. 2009). pH changes were first measured using conventional pH indicators in agar (Marschner and Römheld 1983). This approach has been extended by combining this technique with videodensiometry (Ruiz and Arvieu 1990). More recently optode foils combined with high resolution optical systems have been used for measurement of pH gradients and CO2 release (Blossfeld and Gansert 2007; Rudolph-Mohr et al. 2015). Bioreporters (i.e., bacteria tagged with fluorescent proteins to report a specific activity) have been used to study the release of AsIII (Kuppardt et al. 2010), available nitrate (DeAngelis et al. 2005) and the communication of root colonizing rhizobacteria (Gantner et al. 2006). However, there is clearly merit in doing more work to correlate these bioreporter findings with structural and chemical mapping of the rhizosphere. The extent of phosphorus depletion zones was imaged via autoradiography by Ernst et al. (1989), using 32P and agar media. Recently, Santner et al. (2012) combined diffusive gradients in thin films (DGT) with laser-ablation inductively coupled plasma mass spectroscopy (LA-ICP-MS) to quantify the extent of depletion zones, achieving higher resolutions compared to the older technique.
Future challenges and opportunities
Having provided a brief overview of past work we now discuss the challenges and opportunities for future research in all three areas: structural imaging, chemical mapping and modeling. Our aim is to motivate these somewhat separate communities to work together towards a truly predictive approach to rhizosphere science. We begin with structural and chemical imaging, since without progress in these two domains advances in modeling will be hampered by a lack of data, and the models developed will lack scientific rigor.
Structural imaging using X-ray CT
There are many challenges that the investigator faces when dealing with structural imaging of plant soil interactions. Below we discuss what we consider to be the main ones.
Challenge 1: Image resolution and quality
The spatial and temporal resolution of images obtained via X-ray CT is largely a function of sample material, detector size and the dimensions of the sample within the field of view. Essentially, the finest possible resolution is determined by the size of the object to be scanned; the larger the object, the coarser the resolution. In addition there are numerous hardware concerns associated with high resolution (0.3–1 μm) scanning.
In the ideal case the X-ray flux produced by a benchtop or synchrotron source would be perfectly stable and the detector elements would generate constant output at a given intensity (Ketcham and Carlson 2001). In reality variations in flux, which occur during the scan (Mooney et al. 2012), appear as an apparent shift in attenuation on the radiographs. This manifests in the form of artifacts in the reconstructed data, reducing the ideal correspondence of grey values to X-ray attenuation (Wildenschild and Sheppard 2013). In addition, the scintillation screen (which transforms incident X-ray signal to a visible light signal of proportional intensity) cannot respond instantly to changes in X-ray intensity. This lag time may, if the imaging conditions call for rapid acquisition, result in a given projection being superimposed with the afterglow of the preceding projection (Farukhi 1982). A further concern in benchtop systems is spatial drifting of the spot location on the target. The spot is the region of X-ray emission, and is assumed to be a stable point for the purposes of data reconstruction. Instability in spot size and location during the scan will reduce the certainty with which features in the data can be classified.
A further limitation of detector hardware is the speed with which data can be read from each element. This limit defines the maximal frame rate of the detector (Bigas et al. 2006). In benchtop systems, the exposure times necessitated by the comparatively weak flux (generally > > 50 ms) mean this limit is not exceeded. However, when imaging with high brilliance synchrotron sources, the frame rates for dynamic 4D experiments can be sufficiently high that dedicated high-speed imaging cameras must be used, in conjunction with suitable high efficiency scintillators (i.e., LAG:Ce). Due to the increased scintillator thickness (~100 μm) required for high speed imaging, the spatial resolution is often comparatively poor (>10 μm) (Kalender and Kyriakou 2007).
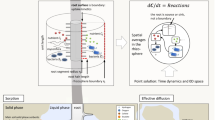
In order to increase the spatial resolution of a scan, it is feasible to image so-called ‘sub-volumes’ within a larger sample, provided that the detector and source can be moved to suitable positions. This does however have implications in terms of pre- and post - filtering of the beam. When features exist in the object that remain outside of the beam for part of or all of the scan, artifacts usually result (Muller and Arce 1996). However, some promising approaches exist for artifact suppression when carrying out region of interest (ROI) tomography, including padding the sinograms of the ROI to a new ‘virtual’ diameter which represents that of the entire object being imaged (see Fig. 2). If this padding is carried out by extending the pixel values at the outer edges of each sinogram, and the number of projections is based not on the ROI diameter, but the diameter of the entire sample, the suppression of artifacts can be highly satisfactory (Kyrieleis et al. 2011).
(a) When imaging an ROI (r1) within a larger sample (r2), the number of radiographs should be calculated based not on the number of pixels across the projection of the ROI (d1) but on the number that would be required to fit a projection of the entire sample at the same magnification (d2). (b) Sinograms should be extended from the diameter of the projected ROI (d1) to the virtual diameter of the projected sample (d2). (c) This padding can be easily achieved by extending the pixel values at the left and right borders out to d2
Another approach to ROI imaging is so-called ‘zoom-in tomography’, in which high-resolution ROI projections are combined with lower-resolution projections of the entire sample in order to suppress artifacts (Xiao et al. 2007). A drawback with this method is that it requires very accurate registration of the two sets of radiographs (Kyrieleis et al. 2011).
Multi-scale imaging is highly important in order to understand the relevance of different scales for mediating key processes occurring in porous media (Cnudde and Boone 2013b). High-resolution data from small subsamples can be registered with lower-resolution volumes which provide information on the macrostructural arrangement of significant features, but it is important to stress that the rhizosphere X-ray dose for the whole sample has to be kept low (<33 Gy) (Zappala et al. 2013b) for the biology not to be affected by the imaging. Thus optimizing the assay size and imaging resolution is of paramount importance, but surprisingly, there are virtually no studies published addressing this specific problem.
Challenge 2: Impact of image quality on interpretation
Surprisingly, little quantitative data is available on how X-ray CT image quality affects subsequent analysis (Schluter et al. 2014). Houston et al. (2013a) and Houston et al. (2013b) proposed methods to assess the quality of images in terms of contrast, noise and sharpness in order to advance our understanding of the impact these parameters have on segmentation of pore space from grey-scale data. They showed that acquisition and reconstruction parameters affect the quality of images, and that this subsequently impacts upon the thresholding outcome. In particular the quality of the image sharpness, controlled by scanning resolution and focus, had a major impact on the thresholding of the data, even when fully automated thresholding algorithms were used (see Fig. 3). Some image analysis is very sensitive to noise (i.e., edge enhancement algorithms and digital volume correlation algorithms), whilst some are particularly sensitive to poor contrast (i.e., histogram-based global segmentation).
Decreasing sharpness increases uncertainty in classification of different phases in CT data. In ideal conditions, the transition between mineral and gas phases should be a step profile (dotted line). In reality, loss of sharpness due to artifacts requires that an interface must be inferred, with the inflection point of the profile often representing the best approximation
One choice researchers must make is to determine between the merits of benchtop and synchrotron X-ray CT systems. Because bench-top systems can now compete with synchrotrons in terms of spatial resolution, one of the major rationales for synchrotron imaging is the comparatively rapid imaging times and the concurrent suppression of motion artifacts when imaging dynamic and/or spatially unstable systems (Wildenschild and Sheppard 2013). Another advantage of synchrotron sources is the monochromatic beam conditions, which allow for phase contrast imaging and absorption edge scanning for element specific analysis, e.g., the use of osmium to visualize SOM. In order to keep energy requirements low and maximize the flux, imaging soils at synchrotron resolutions (~1.5–0.1 μm) presently requires that the sample be in the diameter range of < 5 mm. This raises substantial issues with producing representative samples that can be related to higher scale systems. Recent work by Keyes et al. (2013) has shown that it is possible to grow single roots in soil at a scale amenable to synchrotron imaging. By guiding roots into polymer soil chambers of diameter ~4 mm using rapid-prototyped mesocosms, an intact rhizosphere (albeit for a young plant <3 weeks) can be imaged rapidly enough to suppress motion artifacts visible to the naked eye.
Challenge 3: Identify phases in soil
Though X-ray CT is not a true spectroscopic technique, if a priori information and/or reference data for constituent materials is available (i.e., reference objects imaged using the same parameters), a degree of cautious material identification can be possible for some systems (Cnudde and Boone 2013a). In soils, which typically contain numerous minerals that have different engagement in mechanical deformation, chemical, and flow processes, and often have very similar density and effective atomic number (for example, numerous different forms of silicates), the discernment of specific materials is precluded (Ketcham and Carlson 2001). Nonetheless, tentative distinction between different material classes is often possible (i.e., primary mineral ‘grains’ versus a ‘textural’ phase with a characteristic particle size around or below the imaging resolution), and may be sufficient for some applications, particularly where fluid dynamics rather than soil reactions with plant nutrients are the processes of interest. The nonlinear relationship between X-ray energy and absorption does allow for more refined ‘dual energy’ imaging, taking advantage of abrupt changes in absorption over relatively subtle changes in energy. By imaging a sample at two different mean or peak energies chosen to sit just above and below the K-shell electron binding energy (absorption or ‘k’ edge), distinct image contrast enhancement can be gained for specific materials (Johnson et al. 2007). Though these techniques are widely used in medical radiography, exploration of their application in the geosciences is ongoing (Cnudde 2014). The requirement for a priori knowledge of sample constituents in order to optimize such methods means that if accurate chemical and/or mineralogical discrimination between different phases is required, data fusion of CT data with spectroscopic data from other complementary methods (i.e., XRF/XRD/SEM-EDX/XANES/EXAFS/Raman) is preferable (Hapca et al. 2011).
One cause of non-linearity between material mass attenuation coefficient and actual attenuation coefficient in reconstructed data is the presence of phase effects (Arhatari et al. 2004). As X-rays pass through different material phases, some are absorbed or scattered through atomic interactions, but there is also a velocity change when entering new phases; the velocity being dependent on the density of each material. These velocity shifts result in proportional wavelength changes and shifts in direction (i.e., refraction). The magnitude of these phase differences observed at the detector can be increased simply by adjusting the path-length between the sample and the detector. These effects can be exploited for edge enhancement through simple tuning of the sample to detector distance (in-line propagation phase contrast), and can be very useful for imaging biological low contrast samples. However, this effect can be over-emphasized and cause problems with segmentation in porous media due to the enhancement of gradients in grey-level produced at material boundaries (similar to problems with segmentation of X-ray CT data of porous media due to the partial volume effect). For this reason, it is usually preferable to minimize edge-enhancing phase contrast when applying absorption-domain synchrotron CT imaging to porous media (Wildenschild and Sheppard 2013). However, more sophisticated approaches to phase imaging exist, and are usually synchrotron-based (Stampanoni et al. 2011); though not exclusively (Myers et al. 2007). These methods extract full phase-shift information from the radiographs, which when used for reconstruction can reveal contrast between phases which have very low absorption contrast (Nugent 2010). Wildenschild and Sheppard (2013) identified X-ray phase contrast imaging as a potential technique for imaging biofilms etc. in the soil, but also noted that there are no published studies in this area. This is probably because such imaging is relatively novel, and requires a non-trivial confluence of hardware, technical understanding and mathematical implementation that is not commonly available.
Challenge 4: Quantifying roots at meaningful spatial and temporal scales
There is still a wealth of challenges in developing imaging techniques and protocols for root growth, such as overcoming limitations set by pot size (“bonsai effects”Footnote 3), but also opportunities including the investigation of root growth in structured soils as opposed to the homogenized media used in most studies. In our opinion X-ray CT is still to gain acceptance as a phenotyping tool due to both the comparatively high cost of imaging (as opposed to rhizotron studies and field-based ‘shovelomics’ methods (Trachsel et al. 2011)) and the bottleneck in throughput posed by current image reconstruction and classification protocols. Thus, the question as to whether X-ray CT will remain a research tool best suited for answering specific detailed questions, or also becomes a high-throughput phenotyping tool, depends on the optimization of several processes in this work stream. The image analysis/segmentation tools that do exist for root/rhizosphere research are still in their infancy, and for some applications this may represent the biggest bottleneck. Though some semi-automated root tracking algorithms exist, these have not been successfully or robustly demonstrated for plants significantly more mature than the seedling stage, and they require significant user intervention (Mairhofer et al. 2012). In practice, user-supervised analysis remains a requirement for root segmentation in most scans of larger and more mature plant root architectures (Ahmed et al. 2015; Flavel et al. 2012). The particular strength of X-ray CT imaging currently lies in its suitability for time-resolved imaging of root/soil processes, and correlative or ‘data fusion’ approaches (Ahmed et al. 2015). In the case of fluid flow, it is the one of the few methods, if not the only method, that can reliably provide high spatial- and time-resolution 3D information about processes and structures of relevance in opaque porous media (Wildenschild and Sheppard 2013).
Challenge 5: Unknown effects of X-rays on plants and microbial community
Few studies, especially in the area of plant-soil interaction, report sufficient information to calculate the received radiation dose by the plant. It is known that radiation exposure to seeds during germination impacts growth, with moderate doses (0.01–5 Gy) increasing elongation rates (Johnson 1936). However, larger doses (>15 Gy) inhibit germination and root/shoot elongation rates, observed across a number of species (Genter and Brown 1941; Goodspeed 1929). However exposure after germination appears to be tolerated much better without causing phenotypic change. A study by Zappala et al. (2013a) found no significant differences in major root structure metrics (number of root tips, root volume and root length) or bacterial biomass values between unscanned and repeatedly scanned samples. This was considering an overall dose of ~13 Gy, whereas most individual scans with contemporary bench-top systems produce doses of <1.5 Gy per scan (Zappala et al. 2013a). However, since soil and rhizosphere microbial populations are very diverse, more specific studies investigating how microbes in particular are influenced by X-ray dose are clearly needed. Fischer et al. (2013) have shown that X-ray CT can impact microbial community structure and function significantly, which they explained by the preferential elimination of selected microbial communities through X-ray radiation. However, they also observed a short term recovery of microbial biomass after 7 days and a decrease of differences in the bacterial community structure compared to the situation immediately after scanning. Further they found a clear gradient of effects from the outer to the inner portions of the mesocosm cross-sections, due to the attenuation of X-rays by the soil as the beam traveled through the 20 cm diameter samples (dose was not provided). More detailed studies have been conducted with a total dose in the range of 2.5–7.5 Gy; Bouckaert et al. (2013) and Schmidt et al. (2015) showed no significant impact on any of the key microbial parameters (respiration, enzyme activity, microbial biomass, abundance, community structure) with the exception of archaeal cell numbers. Nevertheless, it is important to determine how repeated scanning influences rhizosphere processes, since one needs to determine a tradeoff between image quality versus the number of scans per lifecycle of the sample. Thus, it is crucial that dose and distribution of dose over time are well documented (Schmidt et al. 2015).
Structural/water imaging using neutron radiography and tomography
Challenge 6: Imaging water distribution around roots
Neutron radiography (see Fig. 4) has been used to study root and water distribution in soils in quasi two-dimensional thin slabs (Menon et al. 2007; Moradi et al. 2009; Oswald et al. 2008). More recently, experiments with time-resolved neutron radiography revealed unexpected water dynamics in the rhizosphere. Carminati et al. (2010) found that the rhizosphere of lupines was wetter than the bulk soil during a drying period, but that following rewetting the rhizosphere remained markedly dry. This dry region extended to 1–2 mm from the root surface. Figure 4 shows radiographs of lupine roots growing in a sandy soil. The sample was dried until a water content of ca. 5 % was reached and then rewetted by capillary rise (the water table was set to a height of 15 cm from the bottom of the sample, while the total soil depth was 30 cm). The radiographs show that the rhizosphere of the upper, older root segments remained dry, while the distal segments of deep roots were quickly rewetted and surrounded by a wet region, probably as a result of mucilage swelling. The details of this experiment can be found in Moradi et al. (2011). These results were confirmed by Carminati (2013) who used neutron tomography to image the rhizosphere in 3D and at a higher spatial resolution. In these experiments the samples (cylinders with diameter of 2.7 cm and height of 10 cm) were scanned in ca. 6 h with a voxel size of 13 μm. Recently, Zarebanadkouki et al. (2015) successfully tomographically imaged samples of the same dimension in 3–6 min with a voxel size of 50 μm.
Neutron radiography of lupine roots in a sandy soil after irrigation by capillary rise (the water table was set at 15 cm depth). The image shows the soil water content θ (red = wet, blue = dry). Roots are visible thanks to their high water content. The sample was 30 cm high, 15 cm large and 1 cm thick. Note the dry rhizosphere around the upper root segments and the wet region around the tips of the deep roots. The figure is modified from Carminati (2013)
Challenge 7: Imaging fluxes in the rhizosphere
Besides imaging water and root distribution, neutron radiation has also been used to quantitatively image fluxes of water into the roots. This process is very important for studying root water uptake. In fact, most studies aiming at predicting the locations of root water uptake have been based on observed changes in soil water content. This approach is motivated by the rationale that root water uptake is higher in more rapidly drying soil regions. Of course, if water redistribution through the soil occurs (and it does), these calculations become more difficult and require the simultaneous modelling of root water uptake and water flow in soils and roots (Javaux et al. 2008). Additionally, the changes in soil water content are more complex than expected and cannot be used to deduce water fluxes across the rhizosphere (Carminati 2012). Therefore, a more direct way to estimate the water fluxes into the root would be of great help for understanding soil-plant water relations.
Matsushima et al. (2008) used neutron radiography to visualize the transport of deuterated water (D2O) in soil and roots. Warren et al. (2013a) and Warren et al. (2013b) used a similar technique to study water redistribution through the root system. Zarebanadkouki et al. (2012) and Zarebanadkouki et al. (2014) developed a method to reconstruct the water fluxes into roots based on D2O injection during both day and night, and simulated the process with an advection–diffusion model of D2O transport into the roots. By inverse simulation of the D2O concentrations in the roots, the authors could reconstruct the water fluxes into a root architecture (Zarebanadkouki et al. 2013). This technique was applied to measure root water uptake in soil regions that were subjected to severe drying and rewetting (Zarebanadkouki and Carminati 2014). The authors found that a drying/wetting cycle temporarily reduced the local uptake of water, probably due to the rhizosphere becoming temporarily hydrophobic after drying.
In conclusion, neutron radiography and tomography are providing new insights into rhizosphere processes and their effect on root water uptake. The higher neutron attenuation of water compared to attenuation by soil particles makes neutron imaging a complementary technique to X-ray CT, where roots are less attenuating than the soil particles.
The same physical effect that is behind the utility of neutron tomography is also one of its limitations. The very high attenuation of neutrons by H2O means that sample sizes are necessarily small in order to reduce the path-lengths through the fluid. The wetter the soil the more pronounced this requirement. This means that the maximum diameter of the sample is in inverse proportion to the water content, and the method is perhaps better suited to the drier end of the possible range of soil water contents observed in field conditions.
The highly penetrating nature of neutrons in many solid materials means that scintillation screens must be substantially thicker than those used in X-ray CT imaging, in order to improve counting statistics and provide workable projection times. This requirement adds uncertainty to the measurement of the attenuated neutrons, since the absorption of the neutron in the scintillator could have occurred anywhere in the through-thickness of the screen. Thus the maximum attainable resolution for neutron imaging is currently at least an order of magnitude poorer than for X-ray CT imaging.
The comparatively low flux of neutron sources (both reactor and spallation) means that scan times have canonically been significantly higher than for X-ray CT imaging. The scan times for neutron tomography are usually in the range of a few hours. However notably, Zarebanadkouki et al. (2015) managed to scan samples in less than 10 min. The relatively long time generally needed for 3D scans limits the ability of neutrons to image water dynamics (doubtless the leading application) and is perhaps the reason why many studies have used 2D transmission imaging rather than tomography, in order not to permit too much averaging of the measured water distributions (see Fig. 4 for illustration). One promising possibility is the use of correlative in situ X-ray CT imaging, using an X-ray beam axis orthogonal to the neutron axis, allowing the high spatial resolution and high mineral contrast of the X-ray imaging to be combined with the excellent H2O mapping capability of the neutron-based approach. With a suitable image registration protocol, these data could be fused for the purpose of parameterizing and/or validating image-based models. The SINQ neutron facility at the Paul Scherrer Institute (Switzerland) now has the functionality for dual neutron/X-ray imaging.
Chemical imaging challenges and opportunities
There is a range of new methods that facilitate chemical imaging with high spatial resolution (nm to μm scale), but always with the tradeoff of small (10 × 10 mm) or very small (10 × 10 μm) sample size. Although the signals used for X-ray, neutron and MRI methods are affected by material properties (electron density, electromagnetic properties, proton content), these methods do not provide true chemical information. The existing methods for chemical speciation are almost all applied to bulk material or 2D thin-sections. One technique with potential for 3D chemical characterization is synchrotron-based μX-ray fluorescence (μXRF) tomography. However, it is currently under debate as to whether self-attenuation of the fluorescence signal will permanently prevent the analysis of samples beyond the millimeter scale (Hapca et al. 2011; Lombi and Susini 2011).
Challenge 8: Non-destructive 3-D chemical mapping
Many chemical imaging techniques are at least partially destructive. They mostly require the exposure of roots and/or soil on a 2D plane, and a number of them also require complete dehydration of the sample as sufficient sensitivity can only be attained under full vacuum conditions. For some techniques, maintenance of spatial arrangement is possible either by embedding the samples with resin or via cryo conservation. This is the case for ToFSIMS, NanoSIMS, μPIXE and SEM-EDX.Footnote 4 These methods provide information on the distribution of elements with higher atomic number (SEM-EDX, μPIXE) or isotopes and fragments of molecules covering all elements (NanoSIMS, ToFSIMS), though for the latter techniques only the ionized fraction is actually detected. An example of chemical mapping of the rhizosphere of a poplar root with ToFSIMS is provided in Martin et al. (2004). Internal structures of the root were visible, as well as the chemical composition of the minerals at the root surface. Further examples for NanoSIMS, a similar technique with much higher resolution, can be found in Clode et al. (2009). While ToFSIMS is able to identify the entire atomic mass range in a single run, NanoSIMS requires that masses of interest be selected prior to the measurement.
Other methods such as Fourier Transform Infrared Spectroscopy (FTIR) and X-ray Photoelectron Spectroscopy (XPS) provide more detailed information on the spatial distribution of different forms of organic carbon in the soil. The former has been applied to map C forms in air-dried thin slices taken from soil micro-aggregates (Lehmann et al. 2007) and was combined with near-edge X-ray absorption fine structure spectroscopy (NEXAFS) for mapping of total organic C content. XPS has the potential to be used for imaging (Barlow et al. 2015). Matrix-assisted laser desorption/ionization mass spectroscopy (MALDI-MS) is another high resolution method, which is able to map the spatial distribution of specific substances including individual effective compounds of pesticides and their metabolites. Currently, MALDI-MS is only applicable to the mapping of root tissue without soil (Rudolph-Mohr et al. 2015).
To take full advantage of these techniques in the rhizosphere, some challenges need to be overcome. Notably the different techniques work at different spatial scales and not all are non-destructive. This necessitates the consideration of protocols that enable integration of physical, chemical and biological techniques. Superimposing different measurements obtained in 2D samples, either thin sections or blocks, is currently possible, but technically challenging. Protocols and software have been developed to start addressing this issue (De Boever et al. 2015; Hapca et al. 2011) or can be borrowed from other disciplines through open access, e.g., Elastix, a software used for registration of medical images (Klein et al. 2010). More recently, Hapca et al. (2015) expanded the work to image registration of multiple planes of SEM-EDX data within a 3D X-ray CT scan and showed how statistical procedures can be used to obtain an estimate of the 3D distribution of chemical elements in soil. Such 3D methods, although presently destructive by nature, are a substantial step forward, allowing integration of physical and chemical techniques to characterize micro-habitats.
Challenge 9: Distinguishing between mobile and immobile phases
None of these new high resolution methods are able to distinguish between the mobile, plant available fraction and the total content of an element. Although, some of them can distinguish different binding forms or functional groups. For investigating mobile forms there are three options based on the use of point sensors/samplers. The first set of approaches carries out solution sampling with small samplers (Puschenreiter et al. 2005; Vetterlein and Jahn 2004); the second utilizes ion-selective electrodes, which have been used successfully to measure ion fluxes in roots (Kochian et al. 1992). Their application for soil based systems is hampered by unsaturated conditions, soil mechanical-impedance and the lack of long term stability in situ. Primarily these electrodes are used to measure NO3 (and K), with a limit of detection approaching 0.1 mM. Unfortunately the ion-selective membranes are sensitive to many other ions including bicarbonate and chloride. The production of a high quality phosphate selective membrane remains a primary challenge (Kim et al. 2009). The third set of approaches uses optodes: 50 μm diameter glass fibers embedded in a metal shaft which can be used as point sensors. The measurement principle is the same as for planar optodes. As optodes can measure in soil, air or soil solution, they address the mobile phases of a particular chemical. Any of these point sensor approaches could theoretically be combined with the 3D non-invasive methods available for visualizing root growth and soil structure, thus providing chemical information in situ in relation to root growth. Hence, these point sensors might provide an alternative approach to addressing the overall challenge, i.e., how to image nutrient concentration profiles at the scale of the rhizosphere, truly in situ, in both time and space. However, these approaches only provide point information and not a complete 3D image.
Garbout et al. (2012) combined positron emission tomography (PET) with X-ray CT to observe the root system of a growing plant. This enabled them to link the observed morphology/structure with imaging of recently assimilated C. The PET scans were used to visualize 11C taken up by the plant through 11C-labelled CO2 and emitted via the root system. Clearly, this is a very promising approach that should be investigated and developed further since some of the issues to do with PET scanning (relatively low spatial resolution) might be resolvable by combining PET with other imaging modalities.
The main challenges for the new emerging field of rhizosphere chemical mapping are: how to correlate structural and chemical data from different imaging modalities, and how to develop sensors that will enable in situ, spatio-temporally resolved monitoring of nutrient (especially phosphate) levels in the soil. Whilst some work exists (in addition to reference above see also Rudolph-Mohr et al. (2014)), we think that the field is still in its infancy, but clearly of crucial importance for rhizosphere science.
Computational and modelling challenges
Challenge 10: Data reconstruction and segmentation
Data reconstruction is an important step in the workstream from sample preparation through imaging, image segmentation and ultimately to image-based models. The suppression of artifacts that hinder image segmentation can be carried out at this stage, when the 3D image volumes are generated from the raw image data. Some of these techniques can be applied prior to reconstruction, including wedge corrections to reduce beam hardening (Ketcham and Carlson 2001), and sinogram correction to suppress ring artifacts. Some artifact mitigation can be carried out on the reconstructed data also, but optimizing for the very best reconstruction is wise since image classification operations can be very time consuming, and become more complex if avoidable artifacts are present.
Reconstruction of parallel-beam (synchrotron) projection data is comparatively simple, since each vertical slice can be considered discretely, and simple filtered back-projection has long been found to give adequate results that are much less computationally intensive than algebraic methods, which require the solution of very large systems of linear equations (Wildenschild and Sheppard 2013). When experiments involve in situ rigs (i.e., a uniaxial compression cell or pressure-plate apparatus), reduced angular access may be an issue due to the presence of lines, cables and other experimental equipment that impede rotation or scatter X-rays (Cnudde and Boone 2013a). In such cases it can be highly beneficial to use iterative algorithms that can produce usable reconstructions when a statistically optimal quantity of projections is unavailable.
The next computational step in the work stream after the image reconstruction is image segmentation, and a good recent review is available in this area by Schluter et al. (2014). A number of papers have recently presented novel image analysis methods, including fully automated and user friendly software, and reviewed methods for segmenting soil samples (Beckers et al. 2014; Hapca et al. 2011; Houston et al. 2013a, b; Iassonov et al. 2009; Kravchenko et al. 2014).
The segmentation of images is the key step in moving away from qualitative assessment of data, and towards quantitative analysis of structures and/or parameterization of mathematical models. In order to carry out such analyses, it is necessary to extract the relevant phases, the definition of which will depend on the subsequent analysis to be carried out. One of the ongoing problems with image segmentation is the wide variance in results that is produced between different datasets and different methods (Baveye et al. 2010).
In porous media research, the discrimination of phases is often divided into gas, fluid and solid. This can sometimes be achieved using global histogram methods, as exhaustively reviewed in Sankur and Sezgin (2001). However, most authors agree that the most pernicious problem in segmenting X-ray CT data of porous media is the partial volume effect. Because the attenuation coefficient of each voxel represents the average of the mass attenuation coefficient for the material system at that location, the presence of porosity at a scale similar to or smaller than the imaging resolution leads to apparent gradients in grey level that are averaged over the voxel volume. Such averaging means that the resulting image does not truly represent the soil structure and may be incorrectly interpreted during segmentation (Cnudde and Boone 2013a).
Because of the existence of partial volume voxels, segmentation approaches must be carefully designed so as not to produce the appearance of erroneous ‘fluid films’ at solid/gas interfaces. Locally adaptive segmentation methods which use local statistical data to refine class assignment are usually found to give better results, as measured using ground-truth datasets generated using phantoms manufactured to a very high tolerance (Schluter et al. 2014). Of these classes of approach, hysteresis segmentation (Vogel and Kretzschmar 1996), watershed imaging (Vincent and Soille 1991), indicator kriging (Oh and Lindquist 1999), later expanded to a fully automated method by (Houston et al. 2013a), Bayesian approaches (Kulkarni et al. 2012) and converging active contours (Sheppard et al. 2004) are among those giving the most satisfactory results (Schluter et al. 2014).
Although scan parameters have a substantial effect on the degree of noise in image data, and some noise can be suppressed during reconstruction, a noise removal step is often applied prior to image segmentation. In the simplest instances, this can be applied using a kernel of a certain size (2D or 3D) which is iteratively centered on different voxel locations, the central voxel having its intensity replaced with some value based on the statistics of the local set of voxels enclosed by the kernel (often the mean or median of the local voxel values). More sophisticated filtering can be achieved by using approximations of physical simulations. A common example of this class of filters is the anisotropic diffusion filter (Perona and Malik 1990), which uses an approximate result of the diffusion equation (i.e., a Gaussian) to preferentially smooth homogenous regions whilst preserving regions of high image gradient (i.e., edges).
Challenge 11: A modelling framework that captures microscopic heterogeneity
Having segmented the images the next step is to implement a modeling framework that uses all the data as efficiently as possible. Image based modelling can be loosely divided into two categories: pore network modelling and direct simulation (Blunt 2001b; Blunt et al. 2013). The first of these, pore network modelling, refers to the extraction of a representative network from the pore scale geometry (Fatt 1956). The pores within the network are assumed to be of a sufficiently simple shape that analytic solutions to the governing equations can be found, reducing the overall computational cost. This method has been widely used to predict averaged transport properties of fluids in beds of packed spheres (Bryant and Blunt 1992, Bryant, King et al. 1993) and imaged porous media (Blunt 2001b; Blunt et al. 2013; Bryant and Blunt 1992). This technique is able to reproduce relative permeability curves and water release characteristics. However, the pore network extraction results in a simplified geometry which may neglect important pore scale phenomena. For example, using pore network models which retain information on pore diameter micro-heterogeneity derived from X-ray CT scans; Perez-Reche et al. (2012) showed that the microscopic heterogeneity ordinarily ignored in most network models has a significant impact on the prediction of soil colonisation by micro-organisms.
The alternative technique of direct modelling involves solving equations directly on the imaged geometries (Raeini et al. 2014b). This technique captures the detail of the pore scale geometry down to the resolution limit. The key disadvantage of direct modeling is that, from a computational point of view, it is highly demanding. Typically a computational mesh has to be generated which conforms to the underlying geometry on which numerical simulations can be run. Mesh generation itself is computationally demanding (Siena et al. 2015) and can be the limiting factor in some simulations. There are numerous methods available for direct solution in the case of single- and multi-phase flow, i.e., finite volume packages such as OpenFOAM (Jasak et al. 2013), ANSYS, FLUENT, and finite element packages such as Comsol Multiphysics, which solve Stokes’ equations directly.
As an alternative to meshes which conform to the geometry, non-structured Cartesian meshes can be designed which use immersed boundary conditions (Mittal and Iaccarino 2005). Immersed boundary conditions make use of a simple mesh of cuboidal elements. The geometrical influence of the boundary is implemented through the addition of source terms in the governing equations which mimic the behavior of the boundary condition. This method has been successfully applied to simulations of flow in simple porous media (Hyman et al. 2012; Siena et al. 2015) and solvers are available which make use of these methods (Prusa et al. 2008).
Direct methods for two-phase fluid flow have a significantly higher computational cost than single phase flow models and are typically solved using Lattice Boltzman methods, as these are highly parallel and relatively easy to implement (Dupuis and Yeomans 2004; Gao et al. 2012; Kusumaatmaja et al. 2006; Kusumaatmaja and Yeomans 2007, 2010; Liu et al. 2014; Ramstad et al. 2010). The Lattice Boltzmann method is slightly different to the more familiar finite volume and finite element methods. Rather than solving a set of partial differential equations, which describe the fluid velocity and pressure at each point in space, a local particle distribution, f i , is defined on a set of discrete lattice points x. The particle distributions are then evolved using an evolution equation
where δt is the time step, ê i is the lattice vector, τ is a relaxation parameter and the function f eq i is chosen such that the evolution equation conserves mass and momentum. The corresponding fluid densities and velocities can be recovered using ρ = ∑ i f i and ρ u = ∑ i f i ê i . Finally the Navier–Stokes equations can be obtained through Taylor expansion of Eq. (1) see, for example Swift et al. (1996) and Zhang et al. (2005).
Whichever the choice of method, there are challenges relating to the discretization and solution of equations. The partial volume effect, mentioned in the previous section, can cause spurious fluid films to be classified at the boundaries between soil particles and pores. This effect is not a problem in highly saturated soils as the majority of flow and transport will occur in the wider pathways. However, as the soil dries the influence of these potentially spurious water films becomes more significant. Hence, the error induced by the partial volume effect can become important. This problem can, of course, be overcome by obtaining X-ray CT scans at higher resolution, effectively pushing the problem to lower saturation values. However, higher resolution comes at the price of increased computational cost, which quickly becomes limiting. Hence, there is a clear need to overcome such limitations through up-scaling methods which use targeted simulations on different scales, effectively minimizing the computational costs of these methods whilst still obtaining sufficient information at each scale. As a way forward, Falconer and Houston (2015) applied a general-purpose-computing-on-graphics-processing-units (GPGPU) approach to a reaction–diffusion soil ecosystem model with the intent of linking the micrometer scale to that of a soil core (cm). They showed that this computational technique can significantly speed up the modelling, and increased the sample size that can be modelled spatially. In addition this could be used to improve visual representation of model outputs.
Challenge 12: Upscaling of processes
There are numerous challenges in the rhizosphere associated with upscaling. Key questions which must be answered are: how does what we see on the microscopic pore-scale (scale of μm) affect the macroscopic properties (e.g., such as the water retention curve, the hydraulic conductivity and the diffusion coefficient) of the rhizosphere (scale of mm)? How can we upscale such rhizosphere findings to the plant/pot scale (scale of 0.1–1 m)? The latter problem should be addressed including the rhizosphere properties (mm-scale) in root architecture models such as those described in a recent review by Dunbabin et al. (2013). However, the first step is to properly define the rhizosphere properties and answer the first question, i.e., how do micro-scale properties affect what we see on the macro-scale, which is the core subject for this review paper.
There are significant mathematical and technical obstacles to be overcome in each of these cases and both rely on being able to accurately upscale from the one scale to another. Due to the complexity of the geometries generated from X-ray CT, access to highly parallel super-computers is essential for Stokes flow calculations (Tracy et al. 2014). Computational cost increases when the full Navier Stokes equations are considered (Icardi et al. 2014); this cost increases further when multiple fluids are simulated. This is in part due to the added non-linearity of fluid interfaces (Anderson et al. 1998) and partly due to the occurrence of thin fluid films which require increased numerical resolution (Raeini et al. 2014a). To give an idea of typical simulation parameters we have calculated the hydraulic conductivity, effective diffusion constant and capillary pressure for cubes of soil ranging in size from 1.78 × 10− 3mm3 to 0.216 mm3. Daly et al. (2015a) have recently shown that, for this well-sieved soil geometry, 0.216 mm3 is sufficient for the effective diffusive properties to converge. Memory usage and simulation time is given in Tables 1 and 2, respectively. Each simulation was carried out on a single 16 core node of the Iridis 4 supercomputing facility at the University of Southampton. Extrapolating these values it was possible to obtain estimates for computational resources required for larger simulations. These quickly become limiting in the case of capillary pressure simulations, with over 60 h simulation time required for a 0.216 mm3 of soil and an estimated 3 weeks simulation time for 1 mm3. However, with ever increasing parallel computational resources, multi-phase flow simulation in porous media is becoming increasingly common (Blunt et al. 2013). Whilst individual simulations on their own offer insight into the flow properties within soil there is a need to go further and make bulk scale predictions based on pore scale simulations. Multiple upscaling techniques exist for this purpose, however, the two most commonly used are volume averaging and homogenization (Hornung 1997).
In this review we focus on homogenization, an area where clear progress towards upscaling has been made. In its simplest form homogenization can be thought of as a formal averaging process. Traditionally this has been carried out for idealized geometries, i.e., close packed spheres or cylinders etc., but the more recent developments involve implementing this method directly on observed (X-ray CT) structures (Daly et al. 2015b; Tracy et al. 2014). The method of homogenization is based on the idea that the underlying porous structure is periodic in some sense, i.e., it is composed of a set of regular repeating units. We consider the example of single phase flow in a porous material (Keller 1980). The porous material has length scale L x and is composed of a set of soil particles with surface Γ and pore space Ω. The pore space is composed of a set of regularly repeating units with period L y as illustrated in Fig. 6. The key assumption used in homogenization is that if \( \epsilon =\frac{L_y}{L_x}\ll 1 \) then we can treat the two length scales independently, i.e., ∇ = ∇ y + ϵ ∇ x and gradients on the scale L x may be considered as a perturbation on the scale L y . We start from Stokes’ equations scaled to L y
where v is the fluid velocity and p is the fluid pressure which we expand as a power series in ϵ,
To proceed we substitute Eq. (3) into Eq. (2) and solve in ascending powers of ϵ. We omit the details, but refer to the books by (Hornung 1997) and (Pavliotis and Stuart 2008). The result is Darcy’s law for u the averaged fluid flow
where ê k is a unit vector in the k -th direction and the local velocity ν k and pressure π w are determined from the cell problem
which is solved on the unit cube. The advantage to this method is that it can be readily applied to images obtained from X-ray CT scanning as illustrated in Figs. 5 and 6 (Tracy et al. 2014).
Homogenization has been successfully used to derive Darcy’s law, and has been applied to single phase flow in single porosity materials (Hornung 1997; Keller 1980) and dual-porosity materials (Arbogast and Lehr 2006; Panfilov 2000). The method has been used to formally derive the Beavers and Joseph condition (Beavers and Joseph 1967) at the interface between a porous material and a free flow region (Jäger and Mikelic 1996; Mikelic and Jäger 2000) and applied to porous media containing voids or vugs (Arbogast and Lehr 2006; Daly and Roose 2014a). In addition, it has been applied to poroelastic media with small deformations (Burridge and Keller 1981) and large deformations (Lee and Mei 1997) and nutrient diffusion processes in soil with root hairs (Leitner et al. 2010a; Zygalakis et al. 2011), nutrient uptake by cluster roots (Zygalakis and Roose 2012) and diffusion of strongly bound nutrients (Ptashnyk and Roose 2010). Multi-fluid homogenization has also been applied to Richards’ equation (Hornung 1997; Panfilov 2000).
Despite the success of this method its use has been mostly restricted to single phase flow when moving from the pore scale to the macro-scale. Dual porosity work has been done to average over several macro-scale solutions for single and dual phase flow (Panfilov 2000). A notable exception is that recently this method has been used to derive a general set of pressure and saturation equations which are shown to reduce to Richards’ equations under an appropriate set of assumptions (Daly and Roose 2015). This method was able to reproduce the water release curve and hydraulic conductivity parameters for simplified geometries, see Fig. 7. There are currently significant computational challenges associated with applying this to images obtained from X-ray CT. Whilst direct methods may work they are computationally expensive and progress may be made by combining these methods with pore network models.
A key difficulty associated with modelling fluid flow on the macroscale is the presence of hysteresis, i.e., the water release curve and the hydraulic conductivity exhibit different values depending on whether the system is undergoing a wetting or a drying cycle (Mualem 1976). Hysteresis in porous media can be loosely classed as having four main causes: the ink-bottle effect which is caused by pore shape, the contact angle hysteresis, compressibility of fluids, and ageing of the soil (Pham et al. 2005). A great deal of work has been carried out on the macroscale modelling of hysteresis, see Albers (2014) and Pham et al. (2005) and references therein. However, relatively little work has been done to determine the relative contribution of each of these effects to the observed hysteresis, although its importance to rhizosphere research is beyond doubt. For example Kroener et al. (2015) demonstrated that mucilage turns hydrophobic upon drying, limiting rhizosphere rewetting.
Challenge 12: Surface roughness
A potential problem can be that resolution limitations and the segmentation methods applied to X-ray-CT data artificially smoothen the solid-pore surface interface (Houston et al. 2013b). Typically larger pores get smoothened and smaller pores are underestimated. There is little work to date that has compared segmentation methods in their prediction of solid-pore interface surface properties, despite the fact that interfaces in soil are critical to the majority of processes and reactions. From a modelling perspective the effect of surface roughness on saturated flow in porous media is well understood. Surface roughness contributes an effective slip length on the surface, a condition widely known as the Beavers and Joseph condition (Beavers and Joseph 1967; Saffman 1971), though alternatives to the Beavers and Joseph condition exist (Levy and Sanchez-Palencia 1975). These conditions have been rigorously justified (Jäger and Mikelic 1996; Mikelic and Jäger 2000) and their contribution to saturated flow has been found (Daly and Roose 2014b). Relatively little work has been done to extend this work to two fluid flow. Mosthaf et al. (2011) use the Beavers and Joseph condition for two fluid flow at the interface between a porous medium and a free flow region, with effective slip length being dependent on saturation. However, they emphasize that this is an approximation and the equivalent Beavers and Joseph condition which accounts for two-phase effects has not yet been derived.
Finally, several issues arise when considering the effect of roots on upscaling. Firstly, a single root will change the properties of soil around it through compaction and excretion of different compounds (Dexter 1987a; Whalley et al. 2005, 2013). These effects cause the soil around the root to vary spatially, an effect that has been studied for the case of a diffusion equation, in which case an advection diffusion equation is derived (Bruna and Chapman 2015). Extending this to consider water movement and upscaling a second time across a set of roots would be an interesting and worthwhile project. Clearly, at some level, possibly on the field scale, this process becomes impossible since natural heterogeneities, such as water courses will start to play a significant role. Hence other techniques will have to be used to account for these features. We will not discuss these methods here since we are aware of the large review currently being processed which covers this issue in great detail (Vereecken et al. 2015).
Conclusions
In this review we have highlighted what we consider to be the key scientific challenges in the area of imaging and predictive modeling of rhizosphere processes. The crucial point, one that we cannot not stress strongly enough, is that model building must go hand in hand with experiments on the structural and chemical properties being considered, otherwise the models will just end up being sophisticated looking “Disney” animations rather than scientifically rigorous and tested models with full predictive power. In our opinion, for the for-seeable future there will not be an “alpha” model for the plant-soil interaction that will be valid for all situations in all environments for all plants. Models will need to be built and calibrated to answer specific scientific questions, and by doing this we will hopefully end up with a library of plant-soil interaction models that will enable the “alpha/beta” model to emerge.
Notes
See the list of terms and abbreviations in the end of the manuscript for detailed definitions of some of the most common terminology and abbreviations.
The partial volume effect is the averaging of attenuation coefficients for materials with features whose characteristic length is below voxel length scale. The result is that a discrete voxel grey level value may actually be encoding an edge between materials with very different attenuation properties.
By “bonsai effect” here we mean the effect of root system crowding due to growth in highly constrained environment not a change due to the wounding of the plant.
See the Terms and Abbreviations list in the end.
Abbreviations
- CLSM:
-
Confocal laser scanning microscopy
- CNR:
-
Contrast to noise ratio
- CT:
-
Computed tomography
- DGT:
-
Diffusive gradient in thin films
- DIC:
-
Digital image correlation
- EXAFS:
-
Extended X-ray absorption fine structure
- FBP:
-
Filtered back projection
- FTIR:
-
Fourier-transform infrared spectroscopy
- GC-MS:
-
Gas chromatography–mass spectrometry
- GPGPU:
-
General-purpose computing on graphics processing units
- HPLC:
-
High-performance liquid chromatography
- HPLC-MS:
-
Liquid chromatography–mass spectrometry
- IR:
-
Infra red
- LA-ICP-MS:
-
Laser ablation inductively coupled plasma mass spectroscopy
- MALDI-MS:
-
Matrix-assisted laser desorption/ionization mass spectroscopy
- MRI:
-
Magnetic resonance imaging
- NanoSIMS:
-
Nano secondary ion mass spectroscopy
- NEXAFS:
-
Near edge X-ray absorption fine structure
- NMR:
-
Nuclear magnetic resonance
- PIV:
-
Particle image velocimetry
- PET:
-
Positron emission tomography
- POM:
-
Particulate organic matter
- Raman:
-
A spectroscopic technique used to observe vibrational rotational, and other low-frequency modes in a system
- ROI:
-
Region of interest
- SEM-EDX:
-
Scanning electron microscope energy dispersive X-ray
- SIRT:
-
Simultaneous iterative reconstruction technique
- SOM:
-
Soil organic matter
- ToFSIMS:
-
Time-of-flight secondary ion mass spectrometry
- XANES:
-
X-ray absorption near edge structure
- XPS:
-
X-ray photoelectron spectroscopy
- X-ray CT:
-
X-ray computed tomography
- XRD:
-
X-ray diffraction
- XRF:
-
X-ray fluorescence
- Zero-sink:
-
Uptake model where the flux of nutrient into the soil is calculated by setting the nutrient concentration at the root surface zero and calculating the resulting flux in soil using the diffusion-convection of it in the soil
- μPIXE:
-
Particle induced X-ray emission or proton-induced X-ray emission
References
Ahmed S, Naugler Klassen T, Keyes SD et al. (2015) Imaging the interaction of roots and phosphate fertiliser granules using 4D X-ray tomography. Plant Soil 2425. doi: 10.1007/s11104-015-2425-5
Albers B (2014) Modeling the hysteretic behavior of the capillary pressure in partially saturated porous media: a review. Acta Mech 1–27
Anderson SH, Hopmans JW (2013) Soil–water–root processes: advances in tomography and imaging, SSSA
Anderson DM, McFadden GB, Wheeler AA (1998) Diffuse-interface methods in fluid mechanics. Annu Rev Fluid Mech 30:139–165. doi:10.1146/Annurev.Fluid.30.1.139
Aravena JE, Berli M, Ghezzehei TA, Tyler SW (2010) Effects of root-induced compaction on rhizosphere hydraulic properties-x-ray microtomography imaging and numerical simulations. Environ Sci Technol 45:425–431
Aravena JE, Berli M, Ghezzehei TA, Tyler SW (2011) Effects of root-induced compaction on rhizosphere hydraulic properties—X-ray microtomography imaging and numerical simulations. Environ Sci Technol 45:425–431. doi:10.1021/es102566j
Aravena J, Berli M, Ruiz S, Suarez F, Ghezzehei T, Tyler S (2014) Quantifying coupled deformation and water flow in the rhizosphere using X-ray microtomography and numerical simulations. Plant Soil 376:95–110. doi:10.1007/s11104-013-1946-z
Arbogast T, Lehr HL (2006) Homogenization of a Darcy–stokes system modeling vuggy porous media. Comput Geosci 10:291–302
Arhatari BD, Mancuso AP, Peele AG, Nugent KA (2004) Phase contrast radiography: Image modeling and optimization. Rev Sci Instrum 75:5271–5276. doi:10.1063/1.1819652
Barber SA (1984) Soil nutrient bioavailability: a mechanistic approach. Wiley-Blackwell
Barlow AJ, Scott O, Sano N, Cumpson PJ (2015) Multivariate Auger Feature Imaging (MAFI)–a new approach towards chemical state identification of novel carbons in XPS imaging. Surf Interface Anal 47(2):173–175
Baveye PC, Laba M, Otten W, Bouckaert L, Dello Sterpaio P, Goswami RR, Grinev D, Houston A, Hu YP, Liu JL, Mooney S, Pajor R, Sleutel S, Tarquis A, Wang W, Wei Q, Sezgin M (2010) Observer-dependent variability of the thresholding step in the quantitative analysis of soil images and X-ray microtomography data. Geoderma 157:51–63. doi:10.1016/J.Geoderma.2010.03.015
Bay BK (2008) Methods and applications of digital volume correlation. J Strain Anal Eng 43:745–760. doi:10.1243/03093247JSA436
Bay BK, Smith TS, Fyhrie DP, Saad M (1999) Digital volume correlation: Three-dimensional strain mapping using X-ray tomography. Exp Mech 39:217–226. doi:10.1007/Bf02323555
Beavers GS, Joseph DD (1967) Boundary conditions at a naturally permeable wall. J Fluid Mech 30:197–207
Beckers E, Plougonven E, Roisin C, Hapca S, Leonard A, Degre A (2014) X-ray microtomography: a porosity-based thresholding method to improve soil pore network characterization? Geoderma 219:145–154. doi:10.1016/J.Geoderma.2014.01.004
Bengough AG (2012) Water dynamics of the root zone: rhizosphere biophysics and its control on soil hydrology. Vadose Zone J 11: doi: 10.2136/vzj2011.0111
Bengough AG, Bransby MF, Hans J, McKenna SJ, Roberts TJ, Valentine TA (2006) Root responses to soil physical conditions; growth dynamics from field to cell. J Exp Bot 57:437–447. doi:10.1093/jxb/erj003
Bengough AG, Hans J, Bransby MF, Valentine TA (2010) PIV as a method for quantifying root cell growth and particle displacement in confocal images. Microsc Res Tech 73:27–36. doi:10.1002/jemt.20749
Bigas M, Cabruja E, Forest J, Salvi J (2006) Review of CMOS image sensors. Microelectron J 37:433–451. doi:10.1016/j.mejo.2005.07.002
Blossfeld S, Gansert D (2007) A novel non‐invasive optical method for quantitative visualization of pH dynamics in the rhizosphere of plants. Plant Cell Environ 30:176–186
Blunt MJ (2001a) Flow in porous media - pore-network models and multiphase flow. Curr Opin Colloid In 6:197–207. doi:10.1016/S1359-0294(01)00084-X
Blunt MJ (2001b) Flow in porous media—pore-network models and multiphase flow. Curr Opin Colloid Interface Sci 6:197–207
Blunt MJ, Bijeljic B, Dong H, Gharbi O, Iglauer S, Mostaghimi P, Paluszny A, Pentland C (2013) Pore-scale imaging and modelling. Adv Water Resour 51:197–216. doi:10.1016/J.Advwatres.2012.03.003
Bouckaert L, Van Loo D, Ameloot N, Buchan D, Van Hoorebeke L, Sleutel S (2013) Compatibility of X-ray micro-computed tomography with soil biological experiments. Soil Biol Biochem 56:10–12. doi:10.1016/j.soilbio.2012.02.002
Brault R, Germaneau A, Dupre JC, Doumalin P, Mistou S, Fazzini M (2013) In-situ analysis of laminated composite materials by X-ray micro-computed tomography and digital volume correlation. Exp Mech 53:1143–1151. doi:10.1007/s11340-013-9730-9
Bruand A, Cousin I, Nicoullaud B, Duval O, Begon JC (1996) Backscattered electron scanning images of soil porosity for analyzing soil compaction around roots. Soil Sci Soc Am J 60:895–901
Bruna M, Chapman SJ (2015) Diffusion in spatially varying porous media. arXiv preprint arXiv:150106549
Bryant S, Blunt M (1992) Prediction of relative permeability in simple porous media. Phys Rev A 46:2004
Burridge R, Keller JB (1981) Poroelasticity equations derived from microstructure. J Acoust Soc Am 70:1140–1146
Carminati A (2012) A Model of root water uptake coupled with Rhizosphere dynamics. Vadose Zone J 11: doi: 10.2136/Vzj2011.0106
Carminati A (2013) Rhizosphere wettability decreases with root age: a problem or a strategy to increase water uptake of young roots? Front Plant Sci 4:298. doi:10.3389/Fpls.2013.00298
Carminati A, Moradi AB, Vetterlein D, Vontobel P, Lehmann E, Weller U, Vogel HJ, Oswald SE (2010) Dynamics of soil water content in the rhizosphere. Plant Soil 332:163–176. doi:10.1007/S11104-010-0283-8
Carminati A, Vetterlein D, Koebernick N, Blaser S, Weller U, Vogel HJ (2013) Do roots mind the gap? Plant Soil 367:651–661. doi:10.1007/s11104-012-1496-9
Clode PL, Kilburn MR, Jones DL, Stockdale EA, Cliff JB, Herrmann AM, Murphy DV (2009) In situ mapping of nutrient uptake in the rhizosphere using nanoscale secondary ion mass spectrometry. Plant Physiol 151:1751–1757
Cnudde V (2014) Introduction: special issue of proceedings of the ‘1st International Conference on Tomography of Materials and Structures’, 1–5 July 2013, Ghent, Belgium, (‘ICTMS2013’). Nucl Instrum Meth B 324:1–3. doi:10.1016/j.nimb.2014.02.005
Cnudde V, Boone M (2013a) High-resolution X-ray computed tomography in geosciences: a review of the current technology and applications. Earth Sci Rev 123:1–17
Cnudde V, Boone MN (2013b) High-resolution X-ray computed tomography in geosciences: a review of the current technology and applications. Earth-Sci Rev 123:1–17. doi:10.1016/j.earscirev.2013.04.003
Daly KR, Roose T (2014a) Multiscale modelling of hydraulic conductivity in vuggy porous media. Proc R Soc A: Math Phys Eng Sci 470: doi: 10.1098/rspa.2013.0383
Daly KR, Roose T (2014a) Multiscale modelling of hydraulic conductivity in vuggy porous media. Proc R Soc A: Math Phys Eng Sci 470:20130383
Daly KR, Roose T (2015) Homogenization of two fluid flow in porous media
Daly KR, Keyes SD, Masum S et al. (2015a) Mage based modeling of nutrient movement in and around the rhizosphere. J Exp Bot (In Press)
Daly KR, Mooney SJ, Bennett MJ, Crout NMJ, Roose T, Tracy SR (2015a) Assessing the influence of the rhizosphere on soil hydraulic properties using X-ray computed tomography and numerical modelling. J Exp Bot 66:2305–2314. doi:10.1093/jxb/eru509
De Boever W, Derluyn H, Van Loo D, Van Hoorebeke L, Cnudde V (2015) Data-fusion of high resolution X-ray CT, SEM and EDS for 3D and pseudo-3D chemical and structural characterization of sandstone. Micrometer 74:15–21. doi:10.1016/j.micrometer.2015.04.003
De Willigen P, Van Noordwijk M (1984) Mathematical-models on diffusion of oxygen to and within plant-roots, with special emphasis on effects of soil-root contact.1. derivation of the models. Plant Soil 77:215–231
De Willigen P, Van Noordwijk M (1994a) Mass-flow and diffusion of nutrients to a root with constant or zero-sink uptake.1. constant uptake. Soil Sci 157:162–170
De Willigen P, Van Noordwijk M (1994b) Mass-flow and diffusion of nutrients to a root with constant or zero-sink uptake.2. zero-sink uptake. Soil Sci 157:171–175
DeAngelis KM, Ji P, Firestone MK, Lindow SE (2005) Two novel bacterial biosensors for detection of nitrate availability in the rhizosphere. Appl Environ Microbiol 71:8537–8547
Dessureault-Rompré J, Nowack B, Schulin R, Luster J (2006) Modified micro suction cup/rhizobox approach for the in-situ detection of organic acids in rhizosphere soil solution. Plant Soil 286:99–107
Dexter AR (1987a) Compression of soil around roots. Plant Soil 97:401–406. doi:10.1007/Bf02383230
Dexter A (1987b) Compression of soil around roots. Plant Soil 97:401–406
Dinkelaker B, Marschner H (1992) In vivo demonstration of acid phosphatase activity in the rhizosphere of soil-grown plants. Plant Soil 144:199–205
Dinkelaker B, Hahn G, Marschner H (1993) Non-destructive methods for demonstrating chemical changes in the rhizosphere II. application of methods. Plant Soil 155:71–74
Downie H, Holden N, Otten W, Spiers AJ, Valentine TA, Dupuy LX (2012) Transparent soil for imaging the rhizosphere. PLoS One 7:e44276. doi:10.1371/journal.pone.0044276
Downie HF, Adu MO, Schmidt S, Otten W, Dupuy LX, White PJ, Valentine TA (2014) Challenges and opportunities for quantifying roots and rhizosphere interactions through imaging and image analysis. Plant Cell Environ. doi:10.1111/pce.12448
Dunbabin VM, Postma JA, Schnepf A, Pages L, Javaux M, Wu LH, Leitner D, Chen YL, Rengel Z, Diggle AJ (2013) Modelling root-soil interactions using three-dimensional models of root growth, architecture and function. Plant Soil 372:93–124. doi:10.1007/S11104-013-1769-Y
Dupuis A, Yeomans JM (2004) Lattice Boltzmann modelling of droplets on chemically heterogeneous surfaces. Futur Gener Comput Syst 20:993–1001
Ernst M, Römheld V, Marschner H (1989) Estimation of phosphorus uptake capacity by different zones of the primary root of soil‐grown maize (Zea mays L.). Z Pflanzenernähr Bodenkd 152:21–25
Falconer RE, Houston AN (2015) Visual simulation of soil-microbial system using GPGPU technology. Compt Rendus Geosci 3:58–71. doi:10.3390/computation3010058
Falconer RE, Battaia G, Schmidt S, Baveye P, Chenu C, Otten W (2015) Microscale heterogeneity explains experimental variability and non-linearity in soil organic matter mineralisation. PLoS One 10:e0123774. doi:10.1371/journal.pone.0123774
Farukhi MR (1982) Recent developments in scintillation detectors for X-ray Ct and Positron Ct applications. IEEE T Nucl Sci 29:1237–1249. doi:10.1109/Tns.1982.4336345
Fatt I (1956) The network model of porous media I. capillary pressure characteristics. Trans AIME 207:144–159
Feeney DS, Crawford JW, Daniell T, Hallett PD, Nunan N, Ritz K, Rivers M, Young IM (2006) Three-dimensional microorganization of the soil-root-microbe system. Microb Ecol 52:151–158. doi:10.1007/s00248-006-9062-8
Fischer D, Pagenkemper S, Nellesen J, Peth S, Horn R, Schloter M (2013) Influence of non-invasive X-ray computed tomography (XRCT) on the microbial community structure and function in soil. J Microbiol Methods 93:121–123. doi:10.1016/j.mimet.2013.02.009
Fitter A (2002) Characteristics and functions of root systems. In: Waisel T, Eshel A, Kafkafi U (eds) Plant roots the hidden half. Marcel Dekker Inc, New York
Flavel RJ, Guppy CN, Tighe M, Watt M, McNeill A, Young IM (2012) Non-destructive quantification of cereal roots in soil using high-resolution X-ray tomography. J Exp Bot 63:2503–2511. doi:10.1093/Jxb/Err421
Forsberg F, Siviour CR (2009) 3D deformation and strain analysis in compacted sugar using x-ray microtomography and digital volume correlation. Meas Sci Technol 20:095703. doi:10.1088/0957-0233/20/9/095703
Forsberg F, Mooser R, Arnold M, Hack E, Wyss P (2008) 3D micro-scale deformations of wood in bending: synchrotron radiation mu CT data analyzed with digital volume correlation. J Struct Biol 164:255–262. doi:10.1016/j.jsb.2008.08.004
Gantner S, Schmid M, Dürr C, Schuhegger R, Steidle A, Hutzler P, Langebartels C, Eberl L, Hartmann A, Dazzo FB (2006) In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol Ecol 56:188–194
Gao W, Ren T, Bengough AG, Auneau L, Watts CW, Whalley WR (2012) Predicting penetrometer resistance from the compression characteristic of soil. Soil Sci Soc Am J 76:361–369. doi:10.2136/sssaj2011.0217
Garbout A, Munkholm LJ, Hansen SB, Petersen BM, Munk OL, Pajor R (2012) The use of PET/CT scanning technique for 3D visualization and quantification of real-time soil/plant interactions. Plant Soil 352:113–127. doi:10.1007/S11104-011-0983-8
Ge Z, Rubio G, Lynch JP (2000) The importance of root gravitropism for inter-root competition and phosphorus acquisition efficiency: results from a geometric simulation model. Plant Soil 218:159–171
Genter CF, Brown HM (1941) X-ray studies on the field bean. J Hered 32:39–44
Ghezzehei TA, Albalasmeh AA (2015) Spatial distribution of rhizodeposits provides built-in water potential gradient in the rhizosphere. Ecol Model 298:53–63. doi:10.1016/j.ecolmodel.2014.10.028
Goodspeed TH (1929) The effects of X-rays and radium on species of genus Nicotana. J Hered 20:243–259
Gregory PJ, Hutchison DJ, Read DB, Jenneson PM, Gilboy WB, Morton EJ (2003) Non-invasive imaging of roots with high resolution X-ray micro-tomography. Plant Soil 255:351–359. doi:10.1023/A:1026179919689
Groenevelt PH, Kay BD, Grant CD (1984) Physical assessment of a soil with respect to rooting potential. Geoderma 34:101–114. doi:10.1016/0016-7061(84)90016-8
Grose MJ, Gilligan CA, Spencer D, Goddard BVD (1996) Spatial heterogeneity of soil water around single roots: use of CT-scanning to predict fungal growth in the rhizosphere. New Phytol 133:261–272. doi:10.1111/J.1469-8137.1996.Tb01893.X
Haase S, Neumann G, Kania A, Kuzyakov Y, Römheld V, Kandeler E (2007) Elevation of atmospheric CO 2 and N-nutritional status modify nodulation, nodule-carbon supply, and root exudation of Phaseolus vulgaris L. Soil Biol Biochem 39:2208–2221
Hall SA, Bornert M, Desrues J, Pannier Y, Lenoir N, Viggiani G, Besuelle P (2010) Discrete and continuum analysis of localised deformation in sand using X-ray mu CT and volumetric digital image correlation. Geotechnique 60:315–322. doi:10.1680/geot.2010.60.5.315
Hapca SM, Wang ZX, Otten W, Wilson C, Baveye PC (2011) Automated statistical method to align 2D chemical maps with 3D X-ray computed micro-tomographic images of soils. Geoderma 164:146–154. doi:10.1016/J.Geoderma.2011.05.018
Hapca S, Baveye P, Wilson C et al. (2015) Three-dimensional mapping of soil chemical characteristics at micrometric scale by combining 2D SEM-EDX data and 3D X-ray. PloS One in press
Helliwell JR, Sturrock CJ, Grayling KM, Tracy SR, Flavel RJ, Young IM, Whalley WR, Mooney SJ (2013) Applications of X-ray computed tomography for examining biophysical interactions and structural development in soil systems: a review. Eur J Soil Sci 64:279–297. doi:10.1111/ejss.12028
Helliwell J, Miller A, Whalley W, Mooney S, Sturrock C (2014a) Quantifying the impact of microbes on soil structural development and behaviour in wet soils. Soil Biol Biochem 74:138–147
Helliwell JR, Miller AJ, Whalley WR, Mooney SJ, Sturrock CJ (2014b) Quantifying the impact of microbes on soil structural development and behaviour in wet soils. Soil Biol Biochem 74:138–147. doi:10.1016/j.soilbio.2014.03.009
Heppell J, Talboys P, Payvandi S, Zygalakis KC, Fliege J, Withers PJA, Jones DL, Roose T (2015) How changing root system architecture can help tackle a reduction in soil phosphate (P) levels for better plant P acquisition. Plant Cell Environ 38:118–128. doi:10.1111/pce.12376
Hinsinger P, Gobran GR, Gregory PJ, Wenzel WW (2005) Rhizosphere geometry and heterogeneity arising from root-mediated physical and chemical processes. New Phytol 168:293–303. doi:10.1111/J.1469-8137.2005.01512.X
Hinsinger P, Bengough AG, Vetterlein D, Young IM (2009) Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil 321:117–152. doi:10.1007/S11104-008-9885-9
Hinsinger P, Brauman A, Devau N, Gerard F, Jourdan C, Laclau JP, Le Cadre E, Jaillard B, Plassard C (2011) Acquisition of phosphorus and other poorly mobile nutrients by roots. where do plant nutrition models fail? Plant Soil 348:29–61. doi:10.1007/S11104-011-0903-Y
Hodge A, Storer K (2015) Arbuscular mycorrhiza and nitrogen: implications for individual plants through to ecosystems. Plant Soil 386:1–19. doi:10.1007/S11104-014-2162-1
Hornung U (1997) Homogenization and porous media. Springer
Houston AN, Schmidt S, Tarquis AM, Otten W, Baveye PC, Hapca SM (2013a) Effect of scanning and image reconstruction settings in X-ray computed microtomography on quality and segmentation of 3D soil images. Geoderma 207:154–165. doi:10.1016/J.Geoderma.2013.05.017
Houston AN, Otten W, Baveye PC, Hapca S (2013b) Adaptive-window indicator kriging: a thresholding method for computed tomography images of porous media. Comput Geosci UK 54:239–248. doi:10.1016/j.cageo.2012.11.016
Huang JY, Pan XC, Li SS, Peng XL, Xiong CY, Fang J (2011) A digital volume correlation technique for 3-D deformation measurements of soft gels. Int J Appl Mech 3:335–354. doi:10.1142/S1758825111001019
Huck MG, Klepper B, Taylor HM (1970) Diurnal variations in root diameter. Plant Physiol 45:529. doi:10.1104/Pp.45.4.529
Hutchings MJ, John EA (2004) The effects of environmental heterogeneity on root growth and root/shoot partitioning. Ann Bot London 94:1–8. doi:10.1093/aob/mch111
Hyman JD, Smolarkiewicz PK, Winter CL (2012) Heterogeneities of flow in stochastically generated porous media. Phys Rev E 86:056701
Iassonov P, Gebrenegus T, Tuller M (2009) Segmentation of X-ray computed tomography images of porous materials: a crucial step for characterization and quantitative analysis of pore structures. Water Resour Res 45:W09415. doi:10.1029/2009wr008087
Icardi M, Boccardo G, Marchisio DL, Tosco T, Sethi R (2014) Pore-scale simulation of fluid flow and solute dispersion in three-dimensional porous media. Phys Rev E 90:013032. doi:10.1103/Physreve.90.013032
Jäger W, Mikelic A (1996) On the boundary conditions at the contact interface between a porous medium and a free fluid. Pisa, Classe Fisiche e Matematiche-Serie IV. Citeseer
Jasak H, Jemcov A, Tukovic Z et al. (2013) OpenFOAM: A C++ library for complex physics simulations
Javaux M, Schroder T, Vanderborght J, Vereecken H (2008) Use of a three-dimensional detailed modeling approach for predicting root water uptake. Vadose Zone J 7:1079–1088. doi:10.2136/vzj2007.0115
Joekar-Niasar V, van Dijke MIJ, Hassanizadeh SM (2012) Pore-scale modeling of multiphase flow and transport: achievements and perspectives. Transp Porous Med 94:461–464. doi:10.1007/S11242-012-0047-4
Johnson EL (1936) Susceptibility of seventy species of flowering plants to X-radiation. Plant Physiol 11:319–342
Johnson TRC, Krauss B, Sedlmair M, Grasruck M, Bruder H, Morhard D, Fink C, Weckbach S, Lenhard M, Schmidt B, Flohr T, Reiser MF, Becker CR (2007) Material differentiation by dual energy CT: initial experience. Eur Radiol 17:1510–1517. doi:10.1007/S00330-006-0517-6
Jones KW, Wang J, Chen Y-C et al. (2013) Tomographic investigations relevant to the rhizosphere. Soil-water-root processes: advances in tomography and imaging SSSA Special Edition 61
Kalender WA, Kyriakou Y (2007) Flat-detector computed tomography (FD-CT). Eur Radiol 17:2767–2779. doi:10.1007/s00330-007-0651-9
Keller JB (1980) Darcy’s law for flow in porous media and the two-space method. Nonlinear partial differential equations in engineering and applied science (Proc Conf, Univ Rhode Island, Kingston, RI, 1979). Dekker New York
Ketcham RA, Carlson WD (2001) Acquisition, optimization and interpretation of X-ray computed tomographic imagery: applications to the geosciences. Comput Geosci 27:381–400
Keyes SD, Daly KR, Gostling NJ, Jones DL, Talboys P, Pinzer BR, Boardman R, Sinclair I, Marchant A, Roose T (2013) High resolution synchrotron imaging of wheat root hairs growing in soil and image based modelling of phosphate uptake. New Phytol 198:1023–1029. doi:10.1111/Nph.12294
Kim HJ, Sudduth KA, Hummel JW (2009) Soil macronutrient sensing for precision agriculture. J Environ Monit 11:1810–1824. doi:10.1039/B906634a
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29:196–205. doi:10.1109/TMI.2009.2035616
Kochian LV, Shaff JE, Kuhtreiber WM, Jaffe LF, Lucas WJ (1992) Use of an extracellular, ion-selective, vibrating microelectrode system for the quantification of K+, H+, and Ca2+ fluxes in maize roots and maize suspension cells. Planta 188:601–610
Koebernick N, Weller U, Huber K et al. (2014) In situ visualization and quantification of three-dimensional root system architecture and growth using X-ray computed tomography. Vadose Zone J 13: doi: 10.2136/vzj2014.03.0024
Koebernick N, Huber K, Kerkhofs E, Vanderborght J, Javaux M, Vereecken H, Vetterlein D (2015) Unraveling the hydrodynamics of split root water uptake experiments using CT scanned root architectures and three dimensional flow simulations. Front Plant Sci 6:370. doi:10.3389/Fpls.2015.00370
Kravchenko AN, Negassa W, Guber AK, Schmidt S (2014) New approach to measure soil particulate organic matter in intact samples using X-ray computed microtomography. Soil Sci Soc Am J 78:1177–1185. doi:10.2136/Sssaj2014.01.0039
Kroener E, Zarebanadkouki M, Kaestner A, Carminati A (2014) Nonequilibrium water dynamics in the rhizosphere: how mucilage affects water flow in soils. Water Resour Res 50:6479–6495. doi:10.1002/2013wr014756
Kroener E, Ahmed MA, Carminati A (2015) Roots at the percolation threshold. Phys Rev E 91:042706. doi:10.1103/Physreve.91.042706
Kulkarni R, Tuller M, Fink W et al. (2012) Three-dimensional multiphase segmentation of X-ray CT data of porous materials using a Bayesian Markov random field framework. Vadose Zone J 11. doi: 10.2136/vzj2011.0082
Kuppardt A, Vetterlein D, Harms H, Chatzinotas A (2010) Visualisation of gradients in arsenic concentrations around individual roots of Zea mays L. using agar-immobilized bioreporter bacteria. Plant Soil 329:295–306
Kusumaatmaja H, Yeomans J (2007) Modeling contact angle hysteresis on chemically patterned and superhydrophobic surfaces. Langmuir 23:6019–6032
Kusumaatmaja H, Yeomans JM (2010) Lattice Boltzmann simulations of wetting and drop dynamics. simulating complex systems by cellular automata. Springer
Kusumaatmaja H, Dupuis A, Yeomans JM (2006) Lattice Boltzmann simulations of drop dynamics. Math Comput Simul 72:160–164
Kyrieleis A, Titarenko V, Ibison M, Connolley T, Withers PJ (2011) Region-of-interest tomography using filtered backprojection: assessing the practical limits. J Microsc (Oxford) 241:69–82. doi:10.1111/J.1365-2818.2010.03408.X
Lee CK, Mei CC (1997) Re-examination of the equations of poroelasticity. Int J Eng Sci 35:329–352
Lehmann J, Kinyangi J, Solomon D (2007) Organic matter stabilization in soil microaggregates: implications from spatial heterogeneity of organic carbon contents and carbon forms. Biogeochemistry 85:45–57
Leitner D, Klepsch S, Ptashnyk M, Marchant A, Kirk G, Schnepf A, Roose T (2010a) A dynamic model of nutrient uptake by root hairs. New Phytol 185:792–802
Leitner D, Schnepf A, Klepsch S, Roose T (2010b) Comparison of nutrient uptake between three‐dimensional simulation and an averaged root system model. Plant Biosyst 144:443–447. doi:10.1080/11263501003726334
Lenoir N, Bornert M, Desrues J, Besuelle P, Viggiani G (2007) Volumetric digital image correlation applied to X-ray microtomography images from triaxial compression tests on argillaceous rock. Strain 43:193–205. doi:10.1111/J.1475-1305.2007.00348.X
Levy T, Sanchez-Palencia E (1975) On boundary conditions for fluid flow in porous media. Int J Eng Sci 13:923–940
Liu H, Kang Q, Leonardi CR et al. (2014) Multiphase lattice Boltzmann simulations for porous media applications--a review. arXiv preprint arXiv:14047523
Lombi E, Susini J (2009) Synchrotron-based techniques for plant and soil science: opportunities, challenges and future perspectives. Plant Soil 320:1–35. doi:10.1007/S11104-008-9876-X
Mairhofer S, Zappala S, Tracy SR, Sturrock C, Bennett M, Mooney SJ, Pridmore T (2012) RooTrak: automated recovery of three-dimensional plant root architecture in soil from X-ray microcomputed tomography images using visual tracking. Plant Physiol 158:561–569. doi:10.1104/Pp.111.186221
Marschner H, Römheld V (1983) In vivo measurement of root-induced pH changes at the soil-root interface: effect of plant species and nitrogen source. Z Pflanzenphysiol 111:241–251
Martin RR, Naftel S, Macfie S, Skinner W, Courchesne F, Séguin V (2004) Time of flight secondary ion mass spectrometry studies of the distribution of metals between the soil, rhizosphere and roots of Populus tremuloides Minchx growing in forest soil. Chemosphere 54:1121–1125
Matsushima U, Kardjilov N, Hilger A, Graf W, Herppich WB (2008) Application potential of cold neutron radiography in plant science research. J Appl Bot Food Qual 82:90–98
McNear DH (2013) The rhizosphere- roots, soil and everything in between. Nat Ed Knowl 4:1
Menon M, Robinson B, Oswald SE, Kaestner A, Abbaspour KC, Lehmann E, Schulin R (2007) Visualization of root growth in heterogeneously contaminated soil using neutron radiography. Eur J Soil Sci 58:802–810. doi:10.1111/j.1365-2389.2006.00870.x
Metzner R, Eggert A, van Dusschoten D, Pflugfelder D, Gerth S, Schurr U, Uhlmann N, Jahnke S (2015) Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: potential and challenges for root trait quantification. Plant Methods 11:17. doi:10.1186/S13007-015-0060-Z
Mikelic A, Jäger W (2000) On the interface boundary condition of Beavers, Joseph, and Saffman. SIAM J Appl Math 60:1111–1127
Mittal R, Iaccarino G (2005) Immersed boundary methods. Annu Rev Fluid Mech 37:239–261
Monga O, Bousso M, Garnier P, Pot V (2008) 3D geometric structures and biological activity: application to microbial soil organic matter decomposition in pore space. Ecol Model 216:291–302. doi:10.1016/j.ecolmodel.2008.04.015
Monga O, Garnier P, Pot V, Coucheney E, Nunan N, Otten W, Chenu C (2014) Simulating microbial degradation of organic matter in a simple porous system using the 3-D diffusion-based model MOSAIC. Biogeosciences 11:2201–2209. doi:10.5194/bg-11-2201-2014
Mooney SJ, Pridmore TP, Helliwell J, Bennett MJ (2012) Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil 352:1–22. doi:10.1007/S11104-011-1039-9
Moradi AB, Conesa HM, Robinson B, Lehmann E, Kuehne G, Kaestner A, Oswald S, Schulin R (2009) Neutron radiography as a tool for revealing root development in soil: capabilities and limitations. Plant Soil 318:243–255. doi:10.1007/s11104-008-9834-7
Moradi AB, Carminati A, Vetterlein D, Vontobel P, Lehmann E, Weller U, Hopmans JW, Vogel HJ, Oswald SE (2011) Three-dimensional visualization and quantification of water content in the rhizosphere. New Phytol 192:653–663. doi:10.1111/J.1469-8137.2011.03826.X
Morgeneyer TF, Helfen L, Mubarak H, Hild F (2013) 3D digital volume correlation of synchrotron radiation laminography images of ductile crack initiation: an initial feasibility study. Exp Mech 53:543–556. doi:10.1007/s11340-012-9660-y
Mosthaf K, Baber K, Flemisch B et al. (2011) A coupling concept for two‐phase compositional porous‐medium and single‐phase compositional free flow. Water Resour Res 47
Mualem Y (1976) Hysteretical models for prediction of the hydraulic conductivity of unsaturated porous media. Water Resour Res 12:1248–1254
Muller M, Arce GR (1996) Truncation artifacts in tomographic reconstructions from projections. Appl Opt 35:3902–3914. doi:10.1364/Ao.35.003902
Myers GR, Gureyev TE, Paganin DM (2007) Stability of phase-contrast tomography. J Opt Soc Am A 24:2516–2526. doi:10.1364/Josaa.24.002516
Neumann G, George TS, Plassard C (2009) Strategies and methods for studying the rhizosphere—the plant science toolbox. Plant Soil 321:431–456
Nugent KA (2010) Coherent methods in the X-ray sciences. Adv Phys 59:1–99. doi:10.1080/00018730903270926
Nunan N, Ritz K, Rivers M, Feeney DS, Young IM (2006a) Investigating microbial micro-habitat structure using X-ray computed tomography. Geoderma 133:398–407
Nunan N, Ritz K, Rivers M, Feeney DS, Young IM (2006b) Investigating microbial micro-habitat structure using X-ray computed tomography. Geoderma 133:398–407. doi:10.1016/j.geoderma.2005.08.004
Nye PH (1994) The effect of root shrinkage on soil-water inflow. Philos T Roy Soc B 345:395–402. doi:10.1098/Rstb.1994.0117
Oh W, Lindquist WB (1999) Image thresholding by indicator kriging. IEEE T Pattern Anal 21:590–602
Olsen SR, Kemper WD (1968) Movemement of nutrients to plant roots. Adv Agron 20:91–151
Oswald SE, Menon M, Carminati A, Vontobel P, Lehmann E, Schulin R (2008) Quantitative imaging of infiltration, root growth, and root water uptake via neutron radiography. Vadose Zone J 7:1035–1047. doi:10.2136/vzj2007.0156
Pagenkemper SK, Peth S, Uteau Puschmann D, Horn R (2013) Effects of root-induced biopores on pore space architecture investigated with industrial X-Ray computed tomography. In: Hopmans JW (ed) Soil-water-root processes: advances in tomography and imaging. SSSA Special Publication. American Society of Agronomy, WI
Pages L (2011) Links between root developmental traits and foraging performance. Plant Cell Environ 34:1749–1760. doi:10.1111/j.1365-3040.2011.02371.x
Pan B, Qian KM, Xie HM, Asundi A (2009) Two-dimensional digital image correlation for in-plane displacement and strain measurement: a review. Meas Sci Technol 20:062001. doi:10.1088/0957-0233/20/6/062001
Panfilov M (2000) Macroscale models of flow through highly heterogeneous porous media. Springer
Pavliotis G, Stuart A (2008) Multiscale methods: averaging and homogenization. Springer Science & Business Media
Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14:399–408. doi:10.1016/j.tplants.2009.05.002
Perez-Reche FJ, Taraskin SN, Otten W, Viana MP, Costa LD, Gilligan CA (2012) Prominent effect of soil network heterogeneity on microbial invasion. Phys Rev Lett 109:098102. doi:10.1103/Physrevlett.109.098102
Perona P, Malik J (1990) Scale-space and edge-detection using anisotropic diffusion. IEEE T Pattern Anal 12:629–639. doi:10.1109/34.56205
Peters WH, Ranson WF (1982) Digital imaging techniques in experimental stress-analysis. Opt Eng 21:427–431. doi:10.1117/12.7972925
Peth S, Nellesen J, Fischer G, Horn R (2010) Non-invasive 3D analysis of local soil deformation under mechanical and hydraulic stresses by mu CT and digital image correlation. Soil Till Res 111:3–18. doi:10.1016/j.sti11.2010.02.007
Peth S, Chenu C, Leblond N, Mordhorst A, Garnier P, Nunan N, Pot V, Ogurreck M, Beckmann F (2014) Localization of soil organic matter in soil aggregates using synchrotron-based X-ray microtomography. Soil Biol Biochem 78:189–194. doi:10.1016/j.soilbio.2014.07.024
Pham HQ, Fredlund DG, Barbour SL (2005) A study of hysteresis models for soil-water characteristic curves. Can Geotech J 42:1548–1568
Pierret A, Doussan C, Capowiez Y, Bastardie F, Pages L (2007) Root functional architecture: a framework for modeling the interplay between roots and soil. Vadose Zone J 6:269–281. doi:10.2136/vzj2006.0067
Prusa JM, Smolarkiewicz PK, Wyszogrodzki AA (2008) EULAG, a computational model for multiscale flows. Comput Fluids 37:1193–1207
Ptashnyk M, Roose T (2010) Derivation of a macroscopic model for transport of strongly sorbed solutes in the soil using homogenization theory. SIAM J Appl Math 70:2097–2118
Puschenreiter M, Wenzel WW, Wieshammer G, Fitz WJ, Wieczorek S, Kanitsar K, Kollensperger G (2005) Novel micro-suction-cup design for sampling soil solution at defined distances from roots. J Plant Nutr Soil Sci-Zeitschrift Fur Pflanzenernahrung Und Bodenkunde 168(3):386–391
Raeini AQ, Blunt MJ, Bijeljic B (2014a) Direct simulations of two-phase flow on micro-CT images of porous media and upscaling of pore-scale forces. Adv Water Resour 74:116–126. doi:10.1016/j.advwatres.2014.08.012
Raeini AQ, Blunt MJ, Bijeljic B et al. (2014b) Direct simulations of two-phase flow on micro-CT images of porous media and upscaling of pore-scale forces. Adv Water Resour
Ramstad T, Oren PE, Bakke S (2010) Simulation of two-phase flow in reservoir rocks using a lattice boltzmann method. SPE J 15:923–933
Roux S, Hild F, Viot P, Bernard D (2008) Three-dimensional image correlation from X-ray computed tomography of solid foam. Compos A Appl Sci 39:1253–1265. doi:10.1016/j.compositesa.2007.11.011
Rudolph-Mohr N, Vontobel P, Oswald SE (2014) A multi-imaging approach to study the root-soil interface. Ann Bot London 114:1779–1787. doi:10.1093/aob/mcu200
Rudolph-Mohr N, Gottfried S, Lamshöft M, Zühlke S, Oswald SE, Spiteller M (2015) Non-invasive imaging techniques to study O 2 micro-patterns around pesticide treated lupine roots. Geoderma 239:257–264
Ruiz L, Arvieu J (1990) Measurement of pH gradients in the rhizosphere. Symbiosis (Rehovot) 9:71–75
Saffman PG (1971) Boundary condition at surface of a porous medium. Stud Appl Math 50:93
Sankur B, Sezgin M (2001) Image thresholding techniques: a survey over categories. Pattern Recogn 34:1573–1607
Santner J, Zhang H, Leitner D, Schnepf A, Prohaska T, Puschenreiter M, Wenzel WW (2012) High-resolution chemical imaging of labile phosphorus in the rhizosphere of Brassica napus L. cultivars. Environ Exp Bot 77:219–226
Schluter S, Sheppard A, Brown K, Wildenschild D (2014) Image processing of multiphase images obtained via X- ray microtomography: a review. Water Resour Res 50:3615–3639. doi:10.1002/2014WR015256
Schmidt S, Bengough AG, Gregory PJ, Grinev DV, Otten W (2012) Estimating root-soil contact from 3D X-ray microtomographs. Eur J Soil Sci 63:776–786. doi:10.1111/j.1365-2389.2012.01487.x
Schmidt H, Vetterlein D, Kohne JM, Eickhorst T (2015) Negligible effect of X-ray mu-CT scanning on archaea and bacteria in an agricultural soil. Soil Biol Biochem 84:21–27. doi:10.1016/j.soilbio.2015.02.010
Sheppard AP, Sok RM, Averdunk H (2004) Techniques for image enhancement and segmentation of tomographic images of porous materials. Physica A 339:145–151. doi:10.1016/J.Physa.2004.03.057
Siena M, Hyman JD, Riva M et al. (2015) Direct numerical simulation of fully saturated flow in natural porous media at the pore scale: a comparison of three computational systems. Comput Geosci 1–15. doi: 10.1007/s10596-015-9486-7
Singh B, Grafe M (2010) Synchrotron-based Techniques in Soils and Sediments. Elsevier, Burlington USA, Amsterdam The Netherlands, Oxford United Kingdom
Spohn M, Kuzyakov Y (2013) Distribution of microbial-and root-derived phosphatase activities in the rhizosphere depending on P availability and C allocation–Coupling soil zymography with 14 C imaging. Soil Biol Biochem 67:106–113
Stampanoni M, Wang ZT, Thuring T, David C, Roessl E, Trippel M, Kubik-Huch RA, Singer G, Hohl MK, Hauser N (2011) The first analysis and clinical evaluation of native breast tissue using differential phase-contrast mammography. Investig Radiol 46:801–806. doi:10.1097/RLI.0b013e31822a585f
Swift MR, Orlandini E, Osborn W, Yeomans J (1996) Lattice Boltzmann simulations of liquid–gas and binary fluid systems. Phys Rev E 54:5041
Timlin D, Ahuja L (2013) Enhancing understanding and quantification of soil–root growth interactions
Tinker PB, Nye PH (2000) Solute movement in the rhizosphere, 2nd edn. Oxford University Press, USA
Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2011) Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341:75–87. doi:10.1007/s11104-010-0623-8
Tracy SR, Daly KR, Sturrock CJ et al. (2014) Three dimensional quantification of soil hydraulic properties using X-ray computed tomography and image based modelling. Water Resour Res
Treseder KK (2013) The extent of mycorrhizal colonization of roots and its influence on plant growth and phosphorus content. Plant Soil 371:1–13. doi:10.1007/S11104-013-1681-5
Van Noordwijk M, De Willigen P (1984) Mathematical-models on diffusion of oxygen to and within plant-roots, with special emphasis on effects of soil-root contact 2. applications. Plant Soil 77:233–241
Vereecken H, Schnepf A, Hopmans J et al. (2015) Modelling soil processes: key challenges and new perspectives. Vadose Zone J submitted Oct 2015
Vetterlein D, Jahn R (2004) Gradients in soil solution composition between bulk soil and rhizosphere–In situ measurement with changing soil water content. Plant Soil 258:307–327
Vincent L, Soille P (1991) Watersheds in digital spaces - an efficient algorithm based on immersion simulations. IEEE T Pattern Anal 13:583–598. doi:10.1109/34.87344
Vogel HJ, Kretzschmar A (1996) Topological characterization of pore space in soil—sample preparation and digital image-processing. Geoderma 73:23–38. doi:10.1016/0016-7061(96)00043-2
Vollsnes AV, Futsaether CM, Bengough AG (2010) Quantifying rhizosphere particle movement around mutant maize roots using time-lapse imaging and particle image velocimetry. Eur J Soil Sci 61:926–939. doi:10.1111/j.1365-2389.2010.01297.x
Warren JM, Bilheux H, Cheng CL, Perfect E (2013a) Reply to: comment on ‘neutron imaging reveals internal plant water dynamics’. Plant Soil 371:15–17. doi:10.1007/s11104-013-1858-y
Warren JM, Bilheux H, Kang MS, Voisin S, Cheng CL, Horita J, Perfect E (2013b) Neutron imaging reveals internal plant water dynamics. Plant Soil 366:683–693. doi:10.1007/s11104-012-1579-7
Whalley WR, Riseley B, Leeds‐Harrison PB, Bird NR, Leech PK, Adderley WP (2005) Structural differences between bulk and rhizosphere soil. Eur J Soil Sci 56:353–360
Whalley W, Ober E, Jenkins M (2013) Measurement of the matric potential of soil water in the rhizosphere. J Exp Bot 64:3951–3963
Wildenschild D, Sheppard AP (2013) X-ray imaging and analysis techniques for quantifying pore-scale structure and processes in subsurface porous medium systems. Adv Water Resour 51:217–246. doi:10.1016/J.Advwatres.2012.07.018
Willert CE, Gharib M (1991) Digital particle image velocimetry. Exp Fluids 10:181–193
Xiao XH, De Carlo F, Stock S (2007) Practical error estimation in zoom-in and truncated tomography reconstructions. Rev Sci Instrum 78:063705. doi:10.1063/1.2744224
Zappala S, Helliwell JR, Tracy SR, Mairhofer S, Sturrock CJ, Pridmore T, Bennett M, Mooney SJ (2013a) Effects of X-ray dose on rhizosphere studies using X-ray computed tomography. PLoS One 8:e67250. doi:10.1371/journal.pone.0067250
Zappala S, Helliwell JR, Tracy SR et al (2013b) Effects of X-ray dose on rhizosphere studies using X-ray computed tomography. Plos One 8. doi: ARTN e67250 10.1371/journal.pone.0067250
Zarebanadkouki M, Carminati A (2014) Reduced root water uptake after drying and rewetting. J Plant Nutr Soil Sci 177:227–236. doi:10.1002/jpln.201300249
Zarebanadkouki M, Kim YX, Moradi AB, Vogel HJ, Kaestner A, Carminati A (2012) Quantification and modeling of local root water uptake using neutron radiography and deuterated water. Vadose Zone J 11:doi: 10.2136/vzj2011.0196
Zarebanadkouki M, Kim YX, Carminati A (2013) Where do roots take up water? neutron radiography of water flow into the roots of transpiring plants growing in soil. New Phytol 199:1034–1044. doi:10.1111/nph.12330
Zarebanadkouki M, Kroener E, Kaestner A, Carminati A (2014) Visualization of root water uptake: quantification of deuterated water transport in roots using neutron radiography and numerical modeling. Plant Physiol 166:487–499. doi:10.1104/pp.114.243212
Zarebanadkouki M, Carminati A, Kestner A et al. (2015) On the fly neutron tomography of water transport into lupine roots. Proceedings of the 10th World Conference on Neutron Radiography (WCNR-10) Grindelwald, Switzerland
Zhang XX, Deeks LK, Bengough AG, Crawford JW, Young LM (2005) Determination of soil hydraulic conductivity with the lattice Boltzmann method and soil thin-section technique. J Hydrol 306:59–70. doi:10.1016/j.jhydrol.2004.08.039
Zygalakis K, Roose T (2012) A mathematical model for investigating the effect of cluster roots on plant nutrient uptake. Eur Phys J Spec Top 204:103–118
Zygalakis K, Kirk G, Jones D, Wissuwa M, Roose T (2011) A dual porosity model of nutrient uptake by root hairs. New Phytol 192:676–688
Acknowledgments
TR would like to acknowledge the receipt of the following funding: Royal Society University Research Fellowship, BBSRC grants BB/C518014, BB/I024283/1, BB/J000868/1, BB/J011460/1, BB/L502625/1, BB/L026058/1, NERC grant NE/L000237/1, EPSRC grant EP/M020355/1 and ERC Consolidator grant 646809 “Data Intensive Modelling of the Rhizosphere Processes”. SDK was funded by EPSRC PhD studentship, BBSRC project BB/J011460/1, EPSRC Doctoral Prize award, and Southampton University Institute for Life Sciences Research Stimulus Fund. KRD is funded by BB/J000868/1. WO would like to acknowledge NERC funding (NE/H01263X/1) and support from the Scottish Alliance for Geoscience, Environment and Society (SAGES). The authors acknowledge the use of the IRIDIS High Performance Computing Facility, and associated support services at the University of Southampton, in the completion of this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Philippe Hinsinger.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Roose, T., Keyes, S.D., Daly, K.R. et al. Challenges in imaging and predictive modeling of rhizosphere processes. Plant Soil 407, 9–38 (2016). https://doi.org/10.1007/s11104-016-2872-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-016-2872-7