Abstract
Key message
Phosphoglycerate Dehydrogenase 1 of the phosphorylated pathway of serine biosynthesis, active in heterotrophic plastids, is required for the synthesis of serine to enable plant growth at high rates of indolic glucosinolate biosynthesis.
Abstract
Plants have evolved effective strategies to defend against various types of pathogens. The synthesis of a multitude of specialized metabolites represents one effective approach to keep plant attackers in check. The synthesis of those defense compounds is cost intensive and requires extensive interaction with primary metabolism. However, how primary metabolism is adjusted to fulfill the requirements of specialized metabolism is still not completely resolved. Here, we studied the role of the phosphorylated pathway of serine biosynthesis (PPSB) for the synthesis of glucosinolates, the main class of defensive compounds in the model plant Arabidopsis thaliana. We show that major genes of the PPSB are co-expressed with genes required for the synthesis of tryptophan, the unique precursor for the formation of indolic glucosinolates (IG). Transcriptional and metabolic characterization of loss-of-function and dominant mutants of ALTERED TRYPTOPHAN1-like transcription factors revealed demand driven activation of PPSB genes by major regulators of IG biosynthesis. Trans-activation of PPSB promoters by ATR1/MYB34 transcription factor in cultured root cells confirmed this finding. The content of IGs were significantly reduced in plants compromised in the PPSB and these plants showed higher sensitivity against treatment with 5-methyl-tryptophan, a characteristic behavior of mutants impaired in IG biosynthesis. We further found that serine produced by the PPSB is required to enable plant growth under conditions of high demand for IG. In addition, PPSB-deficient plants lack the growth promoting effect resulting from interaction with the beneficial root-colonizing fungus Colletotrichum tofieldiae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants produce thousands of different chemical structures classified as specialized metabolites, which are thought to function in facilitating the ecological interaction to optimize plant fitness (Züst and Agrawal 2017). Many of the specialized metabolites are produced in plants to repel specific attackers, but the metabolic cost for the synthesis, modification, transport, maintenance and storage of these compounds is considerable (Gershenzon 1994). Mathematic modeling revealed that maintaining the main defensive trait of plants of the Brassicaceae family, the synthesis of glucosinolates, increases the photosynthetic requirements of the plant by a minimum of 15% (Bekaert et al. 2012). Thus, the biosynthesis of glucosinolates consumes a meaningful percentage of the energy available to the growing plant and requires the development of mechanisms to mitigate the trade-off between growth and the production of defensive compounds.
Glucosinolates are nitrogen and sulfur-rich defense metabolites produced to protect plants against herbivore attack or infection with pathogenic microbes (Halkier and Gershenzon 2006; Hopkins et al. 2009; Pastorczyk and Bednarek 2016). They can be classified into three major groups: aliphatic, indolic and benzenic, depending on their respective amino acid precursor methionine, tryptophan and tyrosine/phenylalanine (Halkier and Gershenzon 2006). Intact glucosinolates are stable and show only little biological activity, while their hydrolysis by myrosinases (β-thioglucsoidases) results in the formation of a vast number of very reactive breakdown products (Wittstock and Burow 2010). Two possible pathways of glucosinolate breakdown have been described (Wittstock et al. 2016). In the “classical” pathway glucosinolates are stored in specialized cells (S-cells) and thus separated from the myrosinase, which is localized in so called myrosine cells. This spatial separation of substrate and enzyme is required to prevent premature glucosinolate breakdown, which occurs only after disruption of the plant tissue by wounding or herbivore attack (Wittstock et al. 2016). In “atypical” glucosinolate breakdown only indolic, but not aliphatic glucosinolates (AG) are hydrolyzed. The degradation of indolic glucosinolates (IG) is catalyzed by the atypical myrosinase PEN2 and occurs within intact cells (Bednarek et al. 2009). This pathway is part of the plant innate immune response and is required to control the entry of filamentous pathogens into the plant (Pastorczyk and Bednarek 2016). Furthermore, recent findings indicate that IG metabolism links plant innate immunity with phosphate nutrition in Arabidopsis plants (Hiruma et al. 2016). While the mineral nutrition of most plant species is supported by interaction with mycorrhizal fungi, around 30% of the plant species, including the Brassicaceae family, are unable to perform this symbiotic interaction (Cosme et al. 2018). However, experimental evidence indicates that other microbes bridge this gap, which has been impressively shown for the beneficial interaction of Arabidopsis plants with the growth promoting endophytic fungus Colletotrichum tofieldiae (C.t.) (Hiruma et al. 2016). The interaction with C.t. enables Arabidopsis plants to improve their growth and phosphate acquisition under phosphate limiting conditions, but requires a functional IG defense pathway (Bednarek et al. 2009; Hiruma et al. 2016). Mutants deficient in major components of this pathway are no longer able to establish a beneficial interaction with C.t. and the fungus behaves like a pathogen in these plants.
IGs are produced from tryptophan and the first reaction, the conversion of tryptophan into indole-3-acetaldoxime (IAOx), is catalyzed by CYP79B2 and CYP79B3, two redundant cytochrome P450 monooxygenase enzymes (Zhao et al. 2002). While single cyp79b2 or cyp79b3 mutations have no major impact on IG content, the respective double mutant is free of IGs (Zhao et al. 2002; Glawischnig et al. 2004). By multiple catalytic reactions IAOx is converted to the first IG, indole-3-ylmethyl glucosinolate (I3M), which can be further modified by hydroxylation and methylation (Pfalz et al. 2011). Hydroxylation of I3M yields in either 4-hydroxy-indole-3-ylmethyl glucosinolate (4HO-I3M) or 1-hydroxy-indole-3-ylmethyl glucosinolate (1HO-I3M), while methylation yields in 4-methoxy-indole-3-ylmethyl glucosinolate (4MO-I3M) or 1-methoxy-indole-3-ylmethyl glucosinolate (1MO-I3M) (Pfalz et al. 20092011).
Previous investigations discovered that IG biosynthesis is regulated by a clade of MYB transcription factors, namely MYB51/HIG1 (HIGH INDOLIC GLUCOSINOLATE 1), MYB122/HIG2 (HIGH INDOLIC GLUCOSINOLATE 2), and MYB34/ATR1 (ALTERED TRYPTOPHAN REGULATION 1) (Celenza et al. 2005; Gigolashvili et al. 2007b; Malitsky et al. 2008; Frerigmann and Gigolashvili 2014). While either their overexpression or dominant mutation increases the expression of all genes involved in IG biosynthesis and results in elevated IG levels, their loss-of-function mutation leads to a down-regulation of IG genes and reduced levels of IGs (Celenza et al. 2005; Gigolashvili et al. 2007b; Malitsky et al. 2008; Frerigmann and Gigolashvili 2014). Intense characterization of MYB34 and MYB51 further revealed their function in regulating the expression of genes of the shikimate and sulfur assimilation pathway, indicating that both transcription factors are responsible for adjusting the activity of primary metabolism to the requirements of IG biosynthesis (Malitsky et al. 2008; Yatusevich et al. 2010). Importantly, identification of tryptophan biosynthetic genes showed that the expression of these genes is induced by environmental stimuli, such as wounding or pathogen infection, and strongly correlated with the expression of genes of the IG pathway (Niyogi and Fink 1992; Niyogi et al. 1993; Zhao et al. 1998). Hence, under conditions of high demand the biosynthesis of tryptophan is boosted beyond the levels necessary for housekeeping functions, such as protein biosynthesis, to enable the synthesis of tryptophan-derived defense metabolites.
The biosynthesis of tryptophan includes six enzymatic reactions and starts with the transfer of the amino group from glutamine to chorismate, the terminal metabolite of the shikimate pathway. This reaction generates anthranilate and pyruvate and is catalyzed by the anthranilate synthase (Niyogi et al. 1993; Poulsen et al. 1993). Via multiple reactions anthranilate is further converted to indole-3-glycerol, the substrate of the tryptophan synthase complex, which consists of an alpha (TSa) and beta (TSb) subunit. The TSa cleaves indole-3-glycerol into indole and glyceraldehyde-3-phosphate, while TSb catalyzes the condensation of indole with serine to form tryptophan (Miles 2001; Weber-Ban et al. 2001). The last reaction indicates that the formation of tryptophan requires an equimolar concentration of serine. Consequently, a secured supply of serine must to be guaranteed under conditions of high tryptophan demand. In plants serine is produced by three pathways; photorespiration (PR), the phosphorylated pathway of serine biosynthesis (PPSB) and the postulated glycerate pathway (Ros et al. 2014), but it is unknown which pathway initially provides serine for tryptophan biosynthesis.
In this study, we set out to identify the mechanism which enables the plant to provide sufficient serine for the formation of tryptophan when the demand for the synthesis of IGs is high. Evidence for at least two distinct serine pools for the formation of tryptophan were previously described (Glawischnig et al. 2000; Benstein et al. 2013). Here, we show that the PPSB is co-activated with tryptophan biosynthesis and this regulation is mediated by the MYB34 transcription factor. In addition, we found that PPSB-derived serine is critical for the trade-off between plant growth and defense.
Results
Genes of the PPSB revealed high expression upon biotic stress and were co-regulated with tryptophan biosynthesis
The analysis of publicly available expression data revealed that the genes of the PPSB, namely PGDH1, PGDH2 and PSAT1 are strongly upregulated after infection of plants with pathogenic bacteria and fungi (Fig. 1a). The infection of plants with the gram-negative bacteria Pseudomonas syringae, the oomycetes Phytophthora infestans and Phytophthora parasitica and the necrotrophic ascomycetes Botrytis cinerea and Sclerotinia sclerotiorum elevated the expression of PGDH1, PGDH2 and PSAT1 of around two to seven-fold.
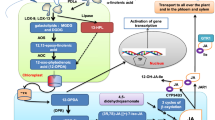
Expression analysis indicated co-regulation of PPSB and tryptophan biosynthetic genes. a Overview on the expression pattern of PPSB and tryptophan biosynthetic genes extracted from publicly available data sources (https://genevestigator.com). Genevestigator ID for the respective experiments are shown in brackets. b Histochemical staining of transgenic plants harboring the PGDH1 promoter ß-galactosidase fusion construct after Mock and methyl jasmonate treatment (MeJA). Bar chart shows PGDH1 expression in seedlings of wild type plants after Mock and MeJA (50 µM) treatment. Data presented are means ± SE of n = 5. Asterisks indicates significantly different values between Mock and MeJA treatment by the Student’s t test (*P < 0.05). c Schematic representation of the biosynthesis of serine, tryptophan, and their relation to indolic glucosinolates, indole-3-acetic acid (IAA), camalexin, and indolecarboxylic acids. The known Arabidopsis thaliana metabolites and proteins involved in these steps are indicated. Metabolites: 3-PGA 3-phosphoglycerate, 3-PHP 3-phosphohydroxypyruvate, 3-PSer 3-phosphoserine, I3GP Indole-3-glycerol phosphate, IAOx indole-3-ylacetaldoxime, I3M indole-3-ylmethyl-glucosinolate, 4HO-I3M 4-hydroxy-indole-3-ylmethyl-glucosinolate, 4MO-I3M 4-methoxy-indole-3-ylmethyl-glucosinolate, 1HO-I3M 1-hydroxy-indole-3-ylmethyl-glucosinolate, 1MO-I3M 1-methoxy-indole-3-ylmethyl-glucosinolate. Multiple enzymatic steps are indicated with interrupted arrows. Proteins. MYB34/MYB51/MYB122 ATR1-like transcription factors 34, 51 and 122, PGDH 3-phosphoglycerate dehydrogenase, PSAT 3-phosphoserine aminotransferase, PSP 3-phosphoserine phosphatase, ASA anthranilate synthase, TSA tryptophan synthase alpha subunit, TSB tryptophan synthase beta subunit, CYP79B2/B3 cytochrome P450 monooxygenase isoform B2 and B3, SUR2 (CYP83B1) cytochrome P450 monooxygenase, GSTF glutathione S-transferase, SUR1 C-S lyase, UGT74B1 UDP-glucose thiohydroximate S-glucosyltransferase, ST5a sulfotransferase, CYP81F2/F3/F4 cytochrome P450 monooxygenase isoform F2, F3 and F4, IGMT1/2 glucosinolate O-methyltransferases isoform 1 and 2, PEN2 beta glucosidase/myrosinase
While biotic stress had a strong influence on the expression of these genes, no substantial changes were observed under various abiotic stresses, except for PGDH2, which was upregulated under osmotic and salt stress conditions (Fig. 1a).
In addition, when analyzing the phytohormone dataset within the Genevestigator Database, we found no strong influence of most phytohormone treatments on the expression of PPSB genes (Fig. 1a). Only the incubation of plants with methyl-jasmonate (MeJA) and salicylic acid (SA), two phytohormones involved in plant defense (Wasternack and Hause 2013; Ding and Ding 2020), elevated the expression of PGDH1, PGDH2 and PSAT1. This finding was validated for PGDH1 expression upon MeJA treatment by RT-qPCR analysis and histological staining of MeJA treated ProPGDH1:GUS lines (Fig. 1b).
Apart from biotic stress, MeJA and SA treatment, the expression of PGDH1, PGDH2 and PSAT1 was also significantly enhanced in plants grown under low phosphate concentrations and after treatment with AgNO3 or UV-B radiation (Fig. 1a).
Interestingly, the expression pattern of PGDH1, PGDH2 and PSAT1 was very similar to the expression of tryptophan biosynthetic genes, such as ASA (anthranilate synthase), IGPS (indole-3-glycerol phosphate synthase), TSA1 (tryptophan synthase alpha subunit) and TRP1 (phosphoribosylanthranilate transferase). The expression of these genes was also elevated in plants after pathogen infection, treatment with MeJA, SA, AgNO3 or UV-B and under phosphate starvation conditions (Fig. 1a). Accordingly, PGDH1, PGDH2 and PSAT1 appear to be in the same gene correlation network as major genes of the tryptophan biosynthesis pathway (Fig. S1).
The comprehensive analysis of the expression of PPSB genes revealed a strong correlation of PGDH1, PGDH2 and PSAT1 expression, while PGDH3, PSAT2 and PSP are differentially expressed (Fig. 1a). The expression of PGDH1, PGDH2 and PSAT1 was high in roots and heterotrophic cells of the leaves, whereas PGDH3 was specifically expressed in mesophyll cells and PSAT2 was expressed only at low levels (Benstein et al. 2013; Toujani et al. 2013; Wulfert and Krueger 2018). PSP is not the rate limiting enzyme of the PPSB and the encoding gene is ubiquitously expressed in the plant (Benstein et al. 2013; Cascales-Minana et al. 2013). Thus, only the major genes of the heterotrophic branch of the PPSB were co-regulated with tryptophan biosynthesis and involved in the response of the plant to the above-mentioned stresses.
Genes of the PPSB are regulated by ATR1/MYB34 transcription factor, a key homeostatic regulator of tryptophan metabolism
To gather more information about the intimate association of PPSB and tryptophan metabolism we investigated the regulation of PPSB genes by ATR1/MYB34 transcription factor (Fig. 2a). Therefore, we analyzed the expression of PGDH1 and PSAT1 in the atr1D dominant mutant (Bender and Fink 1998; Celenza et al. 2005). The enhanced stability of the MYB34 mRNA in the atr1D mutant results in elevated content of MYB34 transcripts (Fig. 2a). Consequently, the transcript levels of the MYB34 target genes, ASA1, TSB1 and CYP79B2 were also significantly higher in the atr1D mutant compared to control plants (Fig. 2a). The analysis of PGDH1 and PSAT1 transcripts revealed substantially elevated levels in atr1D, indicating that also PPSB genes are regulated by the ATR1/MYB34 transcription factor (Fig. 2a).
PPSB genes are regulated by ATR1-like transcription factors. a Expression of MYB34, ASA, TSB, PGDH1, PSAT1 and CYP79B2 in seedlings of atr1D dominant mutant. b Expression of PGDH1, PSAT1 and CYP79B2 in myb-tr (myb34 myb51 myb122) triple mutant. Data presented in (a) and (b) are means ± SE of n = 5. c Trans-activation of PGDH1, PGDH2, PGDH3, PSAT1, PSAT2 and PSP promoter β-glucuronidase (GUS) fusions is shown. Fusion constructs were co-expressed with Pro35S:MYB34 (+) or Pro35S:[empty vector] (−) in cultured Arabidopsis root cells and the GUS activity was determined by staining (upper panel) or measuring the 4-methylumbelliferyl-β-d-glucuronidase activity in nmol (4-MU) min/mg protein (lower panel). Red bar indicates maximum of the y-axis (7 nmol (4-MU) min/mg protein). Data presented are means ± SE of n = 3 (staining) and n = 6 (4-methylumbelliferyl-β-d-glucuronidase activity). Asterisks indicate significantly different values by the Student’s t test (*P < 0.05; **P < 0.01)
In contrast to the elevated transcript level of CYP79B2 gene in the dominant atr1D mutant (Fig. 2a), its expression was strongly reduced in the myb-tr (myb34 myb51 myb122) triple mutant (Fig. 2b). However, the expression of PGDH1 and PSAT1 was not significantly altered in the myb-tr mutant (Fig. 2b), indicating that the ATR1-like transcription factors were not the sole transcriptional regulators of PPSB gene expression.
As mentioned before PGDH1 and PSAT1 expression was substantially up-regulated in the atr1D mutant (Fig. 1a). For a detailed analysis of PPSB gene regulation by ATR1/MYB34 we performed a trans-activation assay using the Arabidopsis root cell culture system (Berger et al. 2007). Staining and fluorometric quantification of β-glucuronidase (GUS) activity showed a significant activation of PGDH1, PGDH2 and PSAT1 promoters by ATR1/MYB34, while the activity of PGDH3, PSAT2 and PSP promoters remained unchanged (Fig. 2c).
In summary, the elevated expression of PGDH1 and PSAT1 in the atr1D mutant and the trans-activation of PGDH1, PGDH2 and PSAT1 promoters by ATR1/MYB34 indicate co-regulation of PPSB with tryptophan-dependent IG biosynthesis.
Over-expression of ATR1/MYB34 transcription factor alters amino acid composition in Arabidopsis plants
Further information about the influence of the ATR1-like transcription factors on the amino acid biosynthesis were obtained by analyzing the amino acid content in myb-tr triple mutants (Fig. 3a) and atr1D mutants (Fig. 3b). The analysis of myb-tr triple mutants revealed no substantial changes in the amino acid content (Fig. 3a). Hence, the three MYB transcription factors were not necessarily required for maintaining amino acid biosynthesis in Arabidopsis plants, supporting its predominant role in plant specialized metabolism (Frerigmann and Gigolashvili 2014).
Elevated indolic glucosinolate biosynthesis alters amino acid contents in plants. The mol percentage of amino acids in seedlings of a myb-tr (myb34 myb51 myb122) triple mutants and of b atr1D single and atr1D cyp79b2/b3 triple mutants is shown. Data presented are means ± SD of n = 5. Letters indicate significantly different values between control (Col-0), atr1D single and atr1D cyp79b2/b3 triple mutant plants by the Student’s t test (P < 0.05)
In contrast to the loss of MYB34/MYB51/MYB122 function, constitutive overexpression of ATR1/MYB34 in the dominant atr1D mutant lead to significant changes in the level of amino acids (Fig. 3b). The levels of asparagine, arginine and lysine increased in atr1D mutants, while the levels of glutamate, serine, glycine, alanine and aspartate decreased. Thus, overexpression of ATR1/MYB34 substantially alters amino acid composition in plants. However, crossing the atr1D mutant with the cyp79b2/b3 (cyp79b2 cyp79b3) double mutant (Celenza et al. 2005) reverted almost all changes in amino acid content (Fig. 3b), indicating that the enhanced metabolic flux into the IG biosynthesis in the atr1D mutant was responsible for the amino acid phenotype. This hypothesis was supported by higher tryptophan content in atr1D cyp79b2/b3 triple mutants (Fig. 3b), which most likely originates from elevated synthesis in combination with impaired turnover into IGs.
Taken together, the ATR1-like clade of transcription factors are not necessarily required to maintain amino acid biosynthesis in Arabidopsis. However, the high demand of amino acid-bound nitrogen for the elevated IG biosynthesis in atr1D mutants causes substantial alterations in amino acid biosynthetic pathways.
A functional PPSB is required for the synthesis of indolic glucosinolates
Analysis of the different glucosinolate species revealed no significant differences in the content of the aliphatic glucosinolates (AG) 3-MSOP (3-(methylsulphinyl)propyl), 4-MSOB (4-(methylsulfinyl)butyl), 5-MSOP (5-(methylsulphinyl)pentyl), 4-MTB (4-(methylthio)butyl) and 8-MSOO (8-(methylsulfinyl)octyl) between Mock-treated PGDH1-silenced lines and empty vector (EV) control plants (Fig. 4). In contrast, PGDH1-silenced lines showed consistent lower levels of the IG I3M (indole-3-ylmethyl) and its processed derivative 1-MO-I3M (1-methoxyindole-3-ylmethyl), while the content of 4-MO-I3M (4-methoxyindole-3-ylmethyl) was not altered (Fig. 4).
PGDH1-silenced lines contain lower amounts of indolic glucosinolates. The concentration of aliphatic (3-MSOP, 4-MSOB, 5-MSOP, 4-MTB, 8-MSOO) and indolic glucosinolates (I3M, 4-MO-I3M, 1-MO-I3M) was determined in seedlings of empty vector control plants (EV) and PGDH1-silenced lines (ts-pgdh1.1, ts-pgdh1.2). Data presented are means ± SD of n = 4. Asterisks indicate significantly different values between EV control plants and PGDH1-silenced lines by the Student’s t test (*P < 0.05)
Most plant pathogens trigger the production of different phytohormones, which are subsequently part of the innate plant immunity (Brader et al. 2001; Kliebenstein et al. 2002; Mikkelsen et al. 2003; Lorenzo et al. 2004; Dombrecht et al. 2007; Hiruma et al. 2010). Some of these hormone signals also impact the biosynthesis of glucosinolates by inducing the expression of main transcriptional regulators (Frerigmann and Gigolashvili 2014). The content of IGs increases strongly after treatment of plants with jasmonate and this response is mainly mediated by the ATR1/MYB34 transcription factor (Frerigmann and Gigolashvili 2014). Therefore, we further investigated the glucosinolate content in Mock and MeJA treated PGDH1-silenced (ts-pgdh1.1, ts-pgdh1.2) and EV (empty vector) control plants (Fig. 4).
While the content of AGs was not significantly different between PGDH1-silenced lines and EV control plants after MeJA treatment, the accumulation of the IG I3M and 1-MO-I3M was significantly reduced (Fig. 4). The content of 4-MO-I3M was slightly, but significantly elevated in PGDH1-silenced lines.
In summary, the analysis of glucosinolates in PGDH1-silenced lines confirmed that the PPSB is required for the synthesis of IGs in Arabidopsis plants.
PPSB-mediated serine biosynthesis is required to maintain plant growth at high levels of indolic glucosinolate biosynthesis
To further study the importance of PPSB-derived serine as precursor for the synthesis of IGs, we generated plants with reduced PGDH1 expression in the background of atr1D mutant plants (ts-pgdh1.1 atr1D; ts-pgdh1.2 atr1D) (Figs. 5 and S2). These plants mimic pathogen-induced activation of IG biosynthesis by simultaneous limitation of PPSB-derived serine. To ensure that the observed phenotypes originate from enhanced flux into IG biosynthesis, we also generated plants additionally lacking the two cytochrome P450 monooxygenase enzymes CYP79B2 and CYP79B3 (ts-pgdh1.1 atr1D cyp79b2/b3; ts-pgdh1.2 atr1D cyp79b2/b3). Both enzymes are essential for the conversion of tryptophan into IAOx (indole-3-acetaldoxime), the precursor for the synthesis of IGs, camalexin and (IAA) indole-3-acetic acid (Mikkelsen et al. 2000; Zhao et al. 2002; Bender and Celenza 2009). The genetic background of all mutants was examined in accordance with previous publications (Zhao et al. 2002; Celenza et al. 2005) and only plants with similar expression values for PGDH1 were selected (Fig. S2).
PGDH1-deficient plants with enhanced indolic glucosinolate biosynthesis are impaired in growth. a Rosette area, shoot and root fresh weight, b primary root growth rate (PRGR), c relative PRGR of plants grown at 5 µM 5-methyl-tryptophan (5-MT) compared to Mock treated plants in percent, d amino acid content in mol percentage, and e glucosinolate content of empty vector (EV) control, PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2), atr1D mutants, ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D double mutants, ts-pgdh1.1 atr1D cyp79b2/b3 and ts-pgdh1.2 atr1D cyp79b2/b3 quadruple mutants, and atr1D cyp79b2/b3 triple mutants are shown. Data presented are means ± SE (a), (b) and (c) n > 20; (d) and (e) n = 5. Different letters (a, b, d, e) indicate significantly different values (P < 0.05). Asterisks (c) indicate significant different values between Mock treated and 5-MT treated plants by the Student’s t test (P < 0.05)
It has been previously described that reducing the activity of PGDH1 in Arabidopsis impairs plant development (Benstein et al. 2013). Similarly, previous characterization of atr1D mutants also revealed a significant impact of the mutation on plant growth performance (Gigolashvili personal communication). Our study confirms the growth phenotype of these transgenic and mutant plants by showing that the rosette area, the shoot and root fresh weight, and the PRGR (primary root growth rate) were significantly reduced in ts-pgdh1.1, ts-pgdh1.2 lines and atr1D mutants (Figs. 5a, b and S2). However, the growth defect of these single mutants was dramatically enhanced in the ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D double mutants (Fig. 5a, b). While the rosette area of ts-pgdh1.1, ts-pgdh1.2 and atr1D lines was reduced to 60 and 40% of control plants, ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D possessed only 10% of the rosette area of control plants. Similarly, the shoot and root fresh weight and the PRGR was substantially lower in ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D double mutants compared to the respective single mutants and EV control plants (Fig. 5a, b). Interestingly, all growth parameters (rosette area, shoot and root fresh weight, PRGR) were significantly improved in ts-pgdh1.1 atr1D cyp79b2/3 and ts-pgdh1.2 atr1D cyp79b2/3 mutants compared to the ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D lines (Fig. 5a, b). Therefore, our data indicated that the enhanced flux of precursors into the IG pathway caused the strong growth phenotype of ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D lines.
Plants impaired in tryptophan metabolism have been described to be more sensitive to treatment with the tryptophan analogue 5-methyl-tryptophan (5-MT) (Bender and Celenza 2009). The toxicity of 5-MT is based on its inhibitory effect on the anthranilate synthase enzyme (Miozzari et al. 1977). Inhibition of anthranilate synthase reduces tryptophan biosynthesis and results in tryptophan starvation. Genetic screens for 5-MT resistance in Arabidopsis revealed several mutants altered in the biosynthesis of tryptophan and IGs (Bender and Celenza 2009). While plants with elevated tryptophan and IG biosynthesis, such as the atr1D mutant are more resistant to 5-MT, plants with reduced IG biosynthesis are more sensitive (Zhao et al. 2002). The latter results from the capacity of Arabidopsis plants to detoxify 5-MT by sequestration into IGs (Zhao et al. 2002). Growth of ts-pgdh1.1 and ts-pgdh1.2 lines at non-toxic (5 µM) 5-MT concentrations reduced the PRGR to 70%, whereas the PRGR of EV control plants was not affected (Fig. 5c). Accordingly, atr1D mutants, which are more resistant to 5-MT (Celenza et al. 2005) had no growth benefit at such low 5-MT concentrations. The analysis of ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D lines also showed no significant impact of 5-MT treatment on the PRGR. This effect is most likely based on the enhanced sequestration of 5-MT into IGs in these plants, which is supported by our finding that the PRGR of ts-pgdh1.1 atr1D cyp79b2/3 and ts-pgdh1.2 atr1D cyp79b2/3 lines grown at 5-MT is reduced to below 60%, and thus on nearly the same level as found for the atr1D cyp79b2/3 mutant (Fig. 5c).
The impact of the different genetic modifications on the development of these mutants indicated that the metabolism of the amino acid precursor and IGs was substantially altered in these plants. This assumption is supported by changes in the level of serine and tryptophan in the different plant genotypes (Fig. 5d). The mol % of serine was significantly lower in ts-pgdh1.1, ts-pgdh1.2 lines and atr1D mutants compared to the EV control plants. This effect was somewhat stronger in ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D lines and mitigated in ts-pgdh1.1 atr1D cyp79b2/3 and ts-pgdh1.2 atr1D cyp79b2/3 lines. Tryptophan was reduced to a similar level in ts-pgdh1.1, ts-pgdh1.2, atr1D, ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D lines, while it accumulated in ts-pgdh1.1 atr1D cyp79b2/3, ts-pgdh1.2 atr1D cyp79b2/3 and atr1D cyp79b2/3 lines (Fig. 5d). The latter effect is explained by simultaneous stimulation of tryptophan biosynthesis due to the ATR1D dominant mutation and blocking of tryptophan conversion into IGs in these plants.
Finally, we analyzed the glucosinolate content in the different genotypes (Fig. 5e). As mentioned before, the contents of all AGs were not altered in ts-pgdh1.1 and ts-pgdh1.2 lines, while the contents of IGs were significantly reduced. The atr1D mutant revealed no changes in the contents of AGs, but accumulated high contents of IGs (Fig. 5e). Thus, our analysis was in line with previous characterizations of the atr1D mutant (Frerigmann and Gigolashvili 2014). The content of IGs also accumulated massively in ts-pgdh1.1 atr1D and ts-pgdh1.2 atr1D lines, but significantly less compared to the atr1D single mutant (Fig. 5e). This finding indicated that ATR1D-driven IG biosynthesis represents a dominant factor, although the synthesis of the serine precursor is limited. The analysis of ts-pgdh1.1 atr1D cyp79b2/3 and ts-pgdh1.2 atr1D cyp79b2/3 lines confirmed the absence of IGs in plants lacking both CYP79B2 and CYP79B3 enzymes as previously reported (Zhao et al. 2002; Frerigmann and Gigolashvili 2014) and did not reveal changes in the contents of AGs (Fig. 5e).
In summary, induction of IG biosynthesis by simultaneous inhibition of PPSB-mediated serine biosynthesis strongly affected plant growth. This phenotype was mitigated when the flux into the IGs was inhibited. Thus, PPSB-derived serine represents a branch point metabolite required for both, plant growth and IG biosynthesis. Latter was supported by the enhanced sensitivity of ts-pgdh1.1 and ts-pgdh1.2 lines against 5-MT.
PPSB-deficient plants lack growth promotion after infection with the beneficial root endophyte Colletotrichum tofieldiae
Previous studies suggested an implication of PPSB and phosphate starvation response of plants, as the transcripts of PGDH1, PGDH2, as well as PSAT1 accumulated strongly in Arabidopsis plants cultured in liquid medium in the absence of phosphate (Pant et al. 2015). More intriguingly, this increase in expression seems to be significantly reduced in plants lacking PHR1, the main transcriptional regulator of the phosphate starvation response (Rubio et al. 2001; Pant et al. 2015). We confirmed this potential linkage under more physiological growth conditions by testing the expression level of PGDH1 within control (EV) and phr1;phl1 mutant plants, grown for five days at phosphate starvation conditions (Fig. S3). Under these conditions PGDH1 transcript levels were increased in shoot and root tissue of control plants in response to low phosphate, but not in the phr1;phl1 mutant background (Fig. S3). Though in relation to transcript regulation of IPS1 (INDUCED BY PHOSPHATE STARVATION1), a well characterized marker of the phosphate starvation response (Franco-Zorrilla et al. 2007), alterations in PGDH1 expression were rather moderate (Fig. S3).
Previous studies showed that the two transcription factors PHR1 and PHL1 are required for the beneficial interaction between Arabidopsis and C.t. under phosphate limiting conditions (Hiruma et al. 2016), suggesting that PHR1/PHL-mediated activation of the PPSB might play a role in this interaction. Analysis of previously published expression data (Hacquard et al. 2016) supported the important role of plant phosphate status in PPSB regulation and showed that C.t. infection itself is not sufficient to activate PPSB gene expression (Fig. S4).
To gather more information about the role of PPSB-mediated serine biosynthesis in the interaction between Arabidopsis and C.t. we infected ts-pgdh1.1 and ts-pgdh1.2 lines with C.t. and monitored the plant growth response (Fig. 6). Therefore, plants were grown on phosphate sufficient (625 µM) or phosphate limiting (100 µM) conditions and in absence (Mock) or presence (C.t.) of the fungus. While shoot fresh weight of EV control plants grown at phosphate sufficient conditions was not altered after infection with C.t., the shoot fresh weight of PGDH1-silenced lines was significantly reduced (Fig. 6). The same tendency was also shown for the root fresh weight, whereby the difference was statistically significant only for the ts-pgdh1.2 line (Fig. 6).
PGDH1-deficient plants show specific impairment of C. tofieldiae-mediated plant growth promotion. Shoot and root fresh weight, and root length of Mock treated and C.t. infected empty vector (EV) control plants and PGDH1-silenced lines (ts-pgdh1.1, ts-pgdh1.2) grown at sufficient phosphate (625 µM Pi) and low phosphate (100 µM Pi) conditions are shown. Data presented are means ± SD of n = 42. Asterisks indicate significantly different values between Mock treated and C.t. infected plants by the Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001)
Infection of EV control plants grown at phosphate sufficient conditions increased the length of the primary root by around twofold from 52 to 98 mm, while the root length of PGDH1-silenced lines was significantly shorter after C.t. infection (Fig. 6).
At phosphate limiting condition C.t. infection increased the shoot biomass of EV control plants from around 10 to 17 mg per plant. However, infection of PGDH1-silenced lines had no substantial influence on the shoot biomass (Fig. 6). Although the root fresh weight of plants grown at phosphate limiting condition was not significantly altered in both, EV control plants and PGDH1-silenced lines, the length of the primary root of EV control plants was significantly longer (Fig. 6). In contrast, the root length of PGDH1-silenced lines was not changed (ts-pgdh1.1) or even reduced (ts-pgdh1.2) after C.t. infection (Fig. 6).
To test whether the growth reduction of PGDH1-silenced lines after C.t. infection was caused by an enhanced susceptibility, we quantified C.t. biomass by determining the amount of fungus DNA using qPCR (Frerigmann et al. 2021). As indicated by the qPCR analysis, the biomass of C.t. increased in plants grown under phosphate limiting conditions (Fig. S5). However, independent of the phosphate status of the plants, no significant difference in the colonialization with C.t. was observed between EV control plants and PGDH1-silenced lines (Fig. S5).
Altogether, we confirmed that the PPSB is activated under phosphate limitation and this regulation requires the transcription factor couple PHR1 and PHL1. In addition, we found that C.t.-mediated plant growth promotion needs a functional PPSB and growth of PGDH1-silenced lines was significantly hampered after C.t.-infection, although these plants were not more susceptible against the fungus.
Discussion
In the last two decades research on the model plant Arabidopsis thaliana has progressively increased our understanding of glucosinolate metabolism in plants (Halkier 2016). Most chemical structures and proteins involved in the synthesis, modification and transport of glucosinolates have been identified (Halkier 2016). In addition, the paradigm about the function of glucosinolates in plant defense has been changed by the realization that, in addition to their function in herbivore defense, they also play an essential role in controlling the invasion of various filamentous pathogens (Pastorczyk and Bednarek 2016). Nevertheless, our knowledge on how primary metabolism is adjusted to the requirements of glucosinolate biosynthesis is still fragmentary. The energy cost for the synthesis of glucosinolates in plants is considerable and thus requires a tight cooperation with primary metabolism (Bekaert et al. 2012). Previously, several primary metabolic pathways, such as sulfur assimilation, methionine biosynthesis and the shikimate pathway have been identified as part of a network co-regulated with glucosinolate biosynthesis (Gigolashvili et al. 2007b, 2007a; Hirai et al. 2007; Malitsky et al. 2008; Yatusevich et al. 2010).
In our study, we identified the phosphorylated pathway of serine biosynthesis as a new component of the regulatory network required for the synthesis of IGs in Arabidopsis thaliana. Expression analyses revealed that major genes of the PPSB, namely PGDH1, PGDH2 and PSAT1, were significantly co-regulated with core genes of the tryptophan biosynthesis pathway (Figs. 1, S1). Tryptophan biosynthesis is up-regulated under high-demand conditions to balance the need for protein biosynthesis with the need for the synthesis of tryptophan-derived secondary metabolites (Niyogi and Fink 1992; Niyogi et al. 1993; Zhao et al. 1998). Several studies showed that this regulation is mediated by the ATR1-like clade of transcription factors (Malitsky et al. 2008; Frerigmann and Gigolashvili 2014). When investigating the impact of ATR1/MYB34 transcription factor on the expression of PPSB genes, we found that the transcript levels of PGDH1 and PSAT1 were elevated in the atr1D dominant mutant (Fig. 2a). Trans-activation of the respective promoter β-glucuronidase fusions by ATR1/MYB34 confirmed this finding (Fig. 2c). Thus, the co-expression of PPSB genes with genes involved in the biosynthesis of tryptophan and IGs is regulated by the same transcription factor.
Interestingly, we found no changes in expression of PPSB genes in loss-of-function myb-tr triple mutants, suggesting that the regulation of these genes by ATR1-like transcription factors is required only under conditions of a high demand for tryptophan or tryptophan-derived metabolites. The analysis of the amino acid content in myb-tr and atr1D mutants supports this assumption (Fig. 3a, b). Even though no substantial alterations were observed in myb-tr mutants, the contents of some amino acids were significantly changed in atr1D mutants (Fig. 3a, b). The most obvious changes were found for serine and asparagine. The content of serine was decreased, whereas the content of asparagine was increased in atr1D mutants. While the reduced level of serine in atr1D mutants confirmed that serine is required for IG biosynthesis, the elevated level of asparagine indicated increased glucosinolate turnover in these plants (Jeschke et al. 2019). Apart from other functions, asparagine is produced in plants during the hydrolysis of organic cyanides, which are glucosinolate breakdown products (Janowitz et al. 2009). Thus, the increased content of asparagine in combination with the reduced content of serine hinted at an elevated flux through the IG biosynthesis and degradation pathways in atr1D mutants. This hypothesis was confirmed by analyzing the amino acid content in atr1D cyp79b2/b3 triple mutants, which are unable to synthesize IGs (Celenza et al. 2005). Almost all observed changes in the amino acid contents in atr1D mutants were reverted in atr1D cyp79b2/b3 triple mutants (Fig. 3b). Thus, we conclude that the elevated synthesis of tryptophan and subsequently IGs in the atr1D mutant induces changes in the amino acid content in these plants and that serine might be a limiting precursor for IG biosynthesis.
When analyzing the contents of glucosinolates in previously characterized PGDH1-silenced lines (Benstein et al. 2013), lower levels of IGs were found, whereas the contents of AGs were unaltered (Fig. 4). Jasmonate treatment, which is known to boost particularly the synthesis of IGs (Mikkelsen et al. 2003), caused higher levels of IGs in both, control plants and PGDH1-silenced lines (Fig. 4). However, the accumulation of the major IGs I3M and 1-MO-I3M was significantly lower in PGDH1-silenced lines compared to the control plants (Fig. 4), providing an additional evidence for a role of the PPSB in IG biosynthesis. Accordingly, PGDH1-silenced lines displayed higher susceptibility against inhibition of the PRGR by 5-MT (Fig. 5c), a characteristic trait of plants impaired in IG biosynthesis (Zhao et al. 2002). In summary, our findings provide evidence that serine generated by the PPSB is crucial for the synthesis of IGs in Arabidopsis.
To further investigate the role of the PPSB for IG biosynthesis, we aimed at continuously boosting the metabolic flux into the IG pathway by simultaneously limiting the availability of PPSB-derived serine. Therefore, we generated atr1D ts-pgdh1 double mutants. We found that the double mutants accumulated high amounts of IGs, nearly similar to the quantities determined in atr1D single mutants (Fig. 5e), but their growth was massively impaired (Fig. 5a, b). This finding indicated that the availability of serine for housekeeping functions, such as protein or nucleotide biosynthesis is limiting in these plants. The reduced serine content in the atr1D ts-pgdh1 double mutants and the reversion of the growth phenotype after the additional blocking of IG formation, achieved by generating atr1D ts-pgdh1 cyp79b2/b3 quadruple mutants, support this conclusion (Fig. 5a-d). In addition, these data are in agreement with previous findings showing that maintenance of high IG levels takes priority over plant growth in trp3-1 and trp2-8 tryptophan-deficient mutants (Müller and Weiler 2000; Brader et al. 2001). Therefore, our data suggest that serine produced by the PPSB represents a branch point metabolite of particular importance for the trade-off between plant growth and IG biosynthesis.
Previous investigations revealed the importance of IG metabolism for controlling the beneficial interaction with the filamentous fungus C.t. under phosphate limiting conditions (Hiruma et al. 2016). To prevent transition of C.t. from beneficial to pathogenic life-style, Arabidopsis plants control C.t. colonization by accumulating glucosinolates under phosphate starvation conditions (Pant et al. 2015; Hiruma et al. 2016). Activation of glucosinolate biosynthesis is triggered in part by C.t. infection itself (Hacquard et al. 2016), but also occurs in uninfected plants in response to phosphate deficiency (Pant et al. 2015). In accordance, we found that the expression of PPSB genes was up-regulated in response to phosphate starvation (Fig. 1) and this regulation requires the PHR1/PHL transcription factor couple (Fig. S3). Furthermore, the analysis of publicly available expression data (Hacquard et al. 2016) indicated that C.t. infection alone had no influence on the regulation of the PPSB genes (Fig. S4), suggesting that PPSB activation is mainly driven by the phosphate status of the plant.
To further test the significance of the PPSB for this mutualistic interaction, we infected PGDH1-silenced plants grown at phosphate sufficient and limiting condition with C.t. (Fig. 6). We found that infected PGDH1-silenced lines lacked the growth promoting effect observed for control plants independent of the phosphate status (Fig. 6). The shoot biomass of PGDH1-silenced lines grown under sufficient phosphate was lower and the length of the primary root was significantly reduced after C.t. infection compared to Mock-treated plants. In addition, the gain in shoot and root biomass observed for control plants grown under phosphate starvation conditions after C.t. infection was missing in PGDH1-silenced lines. However, quantification of fungal growth revealed no significant differences between control plants and PGDH1-silenced lines (Fig. S5). Thus, our data indicate that IG-mediated plant immunity in PGDH1-silenced lines was sufficient to keep C.t. infection in check, but at the cost of plant growth.
Materials and methods
Plant growth
In this study all experiments were conducted with Arabidopsis thaliana plants of the ecotype Columbia (Col-0). Most of the mutants used in this study have been described previously: atr1D single mutant (Bender and Fink 1998); cyp79b2 cyp79b3 (cyp79b2/b3) double and atr1D cyp79b2/b3 triple mutant (Celenza et al. 2005); myb-tr (Frerigmann and Gigolashvili 2014); empty vector (EV) control plants and PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2) (Benstein et al. 2013; Cascales-Minana et al. 2013; Krueger et al. 2017).
The atr1D ts-pgdh1.1 and atr1D ts-pgdh1.2 double mutants, and the atr1D ts-pgdh1.1 cyp79b2/b3 and atr1D ts-pgdh1.2 cyp79b2/b3 quadruple mutants were generated in this study (Fig. S2). Therefore, atr1D single mutants and atr1D cyp79b2/b3 triple mutants were transformed with the PGDH1-silencing construct previously used for the generation of the ts-pgdh1.1 and ts-pgdh1.2 lines (Krueger et al. 2017). Various double and quadruple mutant lines with an expression level of PGDH1 similar to ts-pgdh1.1 and ts-pgdh1.2 were identified by RT-qPCR and two individual double and quadruple mutant lines were selected for further analysis (Fig. S2). Although the atr1D and atr1D cyp79b2/b3 mutants were previously characterized (Bender and Fink 1998; Celenza et al. 2005), we confirmed the presence of the point mutation in the 5’UTR of the coding region of the MYB34 gene and the presence of the T-DNA insertion in the CYP79B2 and CYP79B3 gene in the generated double and quadruple mutant (for details see Figure legends Fig. S2).
For all experiments, Arabidopsis seeds were surface-sterilized using ethanol (2 × 10 min in 70%, 1 × 10 min 100%) and sown on plates containing half-strength Murashige and Skoog basic salt medium (Duchefa). Seeds were stratified two to four days at 4 °C before the plates were transferred to long-day conditions (16 h photoperiod) for germination. Except where stated otherwise, all experimental plants were grown under controlled conditions (16 h photoperiod 20/18 °C, 150 μmol/m−2/s irradiance, 60% humidity) in growth chambers.
Histological staining
To investigate the influence of MeJA treatment on the activity of the PGDH1 gene, previously characterized plants expressing the uidA (ß-glucuronidase) gene under the control of the PGDH1 promoter (Benstein et al. 2013) were grown on half-strength Murashige and Skoog medium in the presence (50 µM MeJA) or absence (Mock) of MeJA (Fig. 1b). The GUS signal was visualized by staining seedlings with X-Gluc for 12 h at 37 °C, followed by 3 h destaining of the chlorophyll in 75% ethanol. GUS-stained seedlings were cleared by incubation in Hoyer’s Solution (Anderson 1954) (Mixture of chloral hydrate:water:glycerol 3.0:0.8:0.2), placed on a glass slide and photographed using a Leica S8AP0 binocular microscope and the respective LAS-EZ (2.1.0) software package.
Expression analysis
Total RNA extraction and RT-qPCR analysis were performed according to Udvardi et al. (2008). The relative quantification of gene expression was conducted using the ∆CT method and the expression values were normalized to ACTIN. Except stated otherwise, two technical and five biological replicates were analyzed. For primer sequences see Table S1.
Trans-activation assay
Cultured Arabidopsis root cells were transformed using the supervirulent Agrobacterium strain LBA4404.pBBR1MCS.virGN54D harboring the constructs for expressing the MYB34 transcription factor (Berger et al. 2007), the anti-silencing protein p19, and the uidA gene under the control of the respective promoters (Fig. 2c). The transformed cells were grown for three to five days and the GUS activity was visualized by adding 100 µL of staining solution (1 mM X-Gluc, 50 mM NaH2PO4, pH 7.1) to 3 ml of the cells and followed by incubation 12 h at 37 °C.
For quantification of the GUS activity, total protein was extracted from 1.5 ml of pelleted root cell culture. The pellet was homogenized and proteins were extracted using 0.5 ml ice-cold extraction buffer (1 mM EDTA, 0.1% v/v Triton X-100, 50 mM NaH2PO4, pH 7.1). After centrifugation for 20 min at 4 °C at 12,000×g 25 µl of the supernatant was mixed with 200 µl MUG solution (22 mg 4-methylumbelliferyl-b-d-glucuronide in 50 ml extraction buffer) and incubated at 37 °C. The formation of 4-methylumbelliferone was continuously monitored fluorometrically (excitation, 340 nm; emission, 465 nm) using a plate reader. The linear slope of the curve was determined and the values were normalized to the protein content within the extract. The total amount of protein in the extract was quantified according to the Bradford (1976) method.
Metabolite analysis
For quantification of amino acids 50 mg of plant material was frozen into liquid nitrogen and homogenized using a ball mill. The amino acids were extracted with 80% ETOH and the extract was diluted 1:10 with water. Twenty microliters of the diluted extract were injected into the HPLC system. Amino acid were separated using a HyperClone 3u ODS (C18)120 150 × 4.6 mm column (Phenomenex) connected to the Dionex UltiMateTM 3000 system (Thermo Fisher Scientific). The HPLC system held a discontinuous flow gradient comprising solvent A (8.8 mM NaPO4, pH 7.5, and 0.2% (v/v) tetrahydrofuran) and increasing proportion of solvent B (18.7 mM NaPO4, pH 7.5, 32.7% (v/v) MeOH and 20.6% (v/v) acetonitrile), at a flow rate of 0.8 ml/min. Quantification was performed by pre-column derivatisation with ortho-phtalaldehyde (OPA; Grace Davison Discovery Science) and subsequently quantified by fluorescence detection of the obtained derivatives (Lindroth and Mopper 1979).
Extraction and quantification of desulpho-glucosinolates was performed as described previously (Gigolashvili et al. 2007b).
Inhibitor treatment
Seeds were germinated on half-strength Murashige and Skoog medium. Four days after germination seedlings were transferred to vertical plates supplemented with or without 5 µM 5-methyltryptophan (5-MT). The root length of the plants was monitored every day and the primary root growth rate was calculated.
Fungal infection
Co-cultivation of C. tofieldiae and A. thaliana was performed as previously described (Frerigmann et al. 2021). Therefore, fungi and plants were co-cultivated on half-strength Murashige and Skoog medium (750 µM MgSO4, 625 or 100 µM KH2PO4, 10.3 mM NH4NO3, 9.4 mM KNO3, 1.5 mM CaCl2, 55 nM CoCl2, 53 nM CuCl2, 50 µM H3BO3, 2.5 µM KI, 50 µM MnCl2, 520 nM Na2MoO4,15 µM ZnCl2,75 µM Fe-EDTA, and 500 µM morpholineethanesulfonic acid-KOH, pH 5.5) with either high Pi (625 µM Pi) or low Pi (100 µM Pi) concentration. The medium was supplemented with 1% nutrient-poor granulated Difco Agar (BD Biosciences). C. tofieldiae was pregrown on PDA media at 25 °C for at least one day and fungal suspension (50 mg/ml) was prepared as previously described (Frerigmann et al. 2021). For co-cultivation, stratified sterile Arabidopsis seeds were resuspended in 250 µl sterile water and 20 µl fungal suspension was added. After washing two times with 1 ml of 20 mM MgCl2, infected seeds were transferred to high or low Pi plates. For each condition four to five replicate plates with seven seeds per plate were prepared. Plates were placed into a phytochamber and plants were grown at short-day conditions (10 h at 21 °C and 14 h at 19 °C).
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Code availability
Not applicable.
References
Anderson LE (1954) Hoyer’s solution as a rapid permanent mounting medium for bryophytes. Bryologist 57:242–244
Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze-Lefert P (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323:101–106
Bekaert M, Edger PP, Hudson CM, Pires JC, Conant GC (2012) Metabolic and evolutionary costs of herbivory defense: systems biology of glucosinolate synthesis. New Phytol 196:596–605
Bender J, Fink GR (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95:5655–5660
Bender J, Celenza JL (2009) Indolic glucosinolates at the crossroads of tryptophan metabolism. Phytochem Rev 8:25–37
Benstein RM, Ludewig K, Wulfert S, Wittek S, Gigolashvili T, Frerigmann H, Gierth M, Flugge UI, Krueger S (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25:5011–5029
Berger B, Stracke R, Yatusevich R, Weisshaar B, Flügge UI, Gigolashvili T (2007) A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J 50:911–916
Brader G, Tas E, Palva ET (2001) Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol 126:849–860
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cascales-Minana B, Munoz-Bertomeu J, Flores-Tornero M, Anoman AD, Pertusa J, Alaiz M, Osorio S, Fernie AR, Segura J, Ros R (2013) The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25:2084–2101
Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J (2005) The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol 137:253–262
Cosme M, Fernández I, Van der Heijden MGA, Pieterse CMJ (2018) Non-mycorrhizal plants: the exceptions that prove the rule. Trends Plant Sci 23:577–587
Ding P, Ding Y (2020) Stories of salicylic acid: a plant defense hormone. Trends Plant Sci 25:549–565
Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, Kazan K (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19:2225–2245
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Frerigmann H, Gigolashvili T (2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7:814–828
Frerigmann H, Piotrowski M, Lemke R, Bednarek P, Schulze-Lefert P (2021) A network of phosphate starvation and immune-related signaling and metabolic pathways controls the interaction between Arabidopsis thaliana and the beneficial fungus Colletotrichum tofieldiae. Mol Plant Microbe Interact, MPMI08200233R.
Gershenzon J (1994) The cost of plant chemical defense against herbivory: a biochemical perspective. Insect-Plant Interact 5:73–105
Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge UI (2007a) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51:247–261
Gigolashvili T, Berger B, Mock HP, Müller C, Weisshaar B, Flügge UI (2007b) The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 50:886–901
Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101:8245–8250
Glawischnig E, Tomas A, Eisenreich W, Spiteller P, Bacher A, Gierl A (2000) Auxin biosynthesis in maize kernels. Plant Physiol 123:1109–1119
Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido-Oter R, Thon MR, Weimann A, Damm U, Dallery JF, Hainaut M, Henrissat B, Lespinet O, Sacristán S, Loren V, van Themaat E, Kemen E, McHardy AC, Schulze-Lefert P, O’Connell RJ (2016) Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun 7:11362
Halkier BA (2016) Chapter one: general introduction to glucosinolates. Adv Bot Res 80:1–14
Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57:303–333
Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, Goda H, Nishizawa OI, Shibata D, Saito K (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104:6478–6483
Hiruma K, Onozawa-Komori M, Takahashi F, Asakura M, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y (2010) Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell 22:2429–2443
Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O’Connell RJ, Schulze-Lefert P (2016) Root endophyte Colletotrichum tofieldiae Confers plant fitness benefits that are phosphate status dependent. Cell 165:464–474
Hopkins RJ, van Dam NM, van Loon JJ (2009) Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu Rev Entomol 54:57–83
Janowitz T, Trompetter I, Piotrowski M (2009) Evolution of nitrilases in glucosinolate-containing plants. Phytochemistry 70:1680–1686
Jeschke V, Weber K, Moore SS, Burow M (2019) Coordination of glucosinolate biosynthesis and turnover under different nutrient conditions. Front Plant Sci 10:1560
Kliebenstein DJ, Figuth A, Mitchell-Olds T (2002) Genetic architecture of plastic methyl jasmonate responses in Arabidopsis thaliana. Genetics 161:1685–1696
Krueger S, Benstein RM, Wulfert S, Anoman AD, Flores-Tornero M, Ros R (2017) Studying the Function Of The Phosphorylated Pathway Of Serine Biosynthesis in Arabidopsis thaliana. Methods Mol Biol 1653:227–242
Lindroth P, Mopper K (1979) High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with o-phthaldialdehyde. Anal Chem 51:1667–1674
Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16:1938–1950
Malitsky S, Blum E, Less H, Venger I, Elbaz M, Morin S, Eshed Y, Aharoni A (2008) The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol 148:2021–2049
Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem 275:33712–33717
Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131:298–308
Miles EW (2001) Tryptophan synthase: a multienzyme complex with an intramolecular tunnel. Chem Rec 1:140–151
Miozzari G, Niederberger P, Hütter R (1977) Action of tryptophan analogues in Saccharomyces cerevisiae. Arch Microbiol 115:307–316
Müller A, Weiler EW (2000) Indolic constituents and indole-3-acetic acid biosynthesis in the wild-type and a tryptophan auxotroph mutant of Arabidopsis thaliana. Planta 211:855–863
Niyogi KK, Fink GR (1992) Two anthranilate synthase genes in Arabidopsis: defense-related regulation of the tryptophan pathway. Plant Cell 4:721–733
Niyogi KK, Last RL, Fink GR, Keith B (1993) Suppressors of trp1 fluorescence identify a new arabidopsis gene, TRP4, encoding the anthranilate synthase beta subunit. Plant Cell 5:1011–1027
Pant BD, Pant P, Erban A, Huhman D, Kopka J, Scheible WR (2015) Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ 38:172–187
Pastorczyk M, Bednarek P (2016) The function of glucosinolates and related metabolites in plant innate immunity. Adv Bot Res 80:171–181
Pfalz M, Vogel H, Kroymann J (2009) The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21:985–999
Pfalz M, Mikkelsen MD, Bednarek P, Olsen CE, Halkier BA, Kroymann J (2011) Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23:716–729
Poulsen C, Bongaerts RJ, Verpoorte R (1993) Purification and characterization of anthranilate synthase from Catharanthus roseus. Eur J Biochem 212:431–440
Ros R, Munoz-Bertomeu J, Krueger S (2014) Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci 19:564–569
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Toujani W, Munoz-Bertomeu J, Flores-Tornero M, Rosa-Tellez S, Anoman AD, Alseekh S, Fernie AR, Ros R (2013) Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol 163:1164–1178
Udvardi MK, Czechowski T, Scheible WR (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20:1736–1737
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development: an update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058
Weber-Ban E, Hur O, Bagwell C, Banik U, Yang LH, Miles EW, Dunn MF (2001) Investigation of allosteric linkages in the regulation of tryptophan synthase: the roles of salt bridges and monovalent cations probed by site-directed mutation, optical spectroscopy, and kinetics. Biochemistry 40:3497–3511
Wittstock U, Burow M (2010) Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book 8:e0134
Wittstock U, Kurzbach E, Herfurth AM, Stauber EJ (2016) Chapter six: glucosinolate breakdown. Adv Bot Res 80:125–169
Wulfert S, Krueger S (2018) Phosphoserine aminotransferase1 is part of the phosphorylated pathways for serine biosynthesis and essential for light and sugar-dependent growth promotion. Front Plant Sci 9:1712
Yatusevich R, Mugford SG, Matthewman C, Gigolashvili T, Frerigmann H, Delaney S, Koprivova A, Flügge UI, Kopriva S (2010) Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J 62:1–11
Zhao J, Williams CC, Last RL (1998) Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 10:359–370
Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16:3100–3112
Züst T, Agrawal AA (2017) Trade-offs between plant growth and defense against insect herbivory: an emerging mechanistic synthesis. Annu Rev Plant Biol 68:513–534
Funding
Open Access funding enabled and organized by Projekt DEAL. S.K. was funded by the Deutsche Forschungsgemeinschaft (Kr4245/2-1). The research of S.E.Z. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany´s Excellence Strategy EXC-1028 and EXC-2048/1 project ID 390686111.
Author information
Authors and Affiliations
Contributions
SK designed research, performed initial experiments, analyzed data, and wrote the manuscript. SEZ, SB performed plant experiments and analyzed data. HF carried out all microbial assays, and assisted in writing. All author assisted in editing the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there is no conflict of interest.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zimmermann, S.E., Blau, S., Frerigmann, H. et al. The phosphorylated pathway of serine biosynthesis is crucial for indolic glucosinolate biosynthesis and plant growth promotion conferred by the root endophyte Colletotrichum tofieldiae. Plant Mol Biol 107, 85–100 (2021). https://doi.org/10.1007/s11103-021-01181-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01181-5