Abstract
Objective
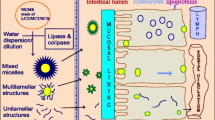
Molecular dynamics (MD) simulations provide an in silico method to study the structure of lipid-based formulations (LBFs) and the incorporation of poorly water-soluble drugs within such formulations. In order to validate the ability of MD to effectively model the properties of LBFs, this work investigates the well-known cyclosporine A formulations, Sandimmune® and Neoral®. Sandimmune® exhibits poor dispersibility and its absorption from the gastrointestinal tract is enhanced when administered after food, whereas Neoral® disperses comparatively well and shows no food effect.
Methods
MD simulations were performed of both LBFs to investigate the differences observed in fasted and fed conditions. These conditions were also tested using an in vitro experimental model of dispersion and digestion.
Results
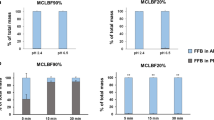
These MD simulations were able to show that the food effect observed for Sandimmune® can be explained by large changes in drug solubilization on addition of bile. In contrast, Neoral® is well dispersed in water or in simulated fasted conditions, and this dispersion is relatively unchanged on moving to fed conditions. These differences were confirmed using dispersion and digestion in vitro experimental model.
Conclusions
The current data suggests that MD simulations are a potential method to model the fate of LBFs in the gastrointestinal tract, predict their dispersion and digestion, investigate behaviour of APIs within the formulations, and provide insights into the clinical performance of LBFs.
Graphical abstract










Similar content being viewed by others
References
Williams HD, Trevaskis NL, Charman SA, Shanker RM, Charman WN, Pouton CW, Porter CJH. Strategies to address low drug solubility in discovery and development. Pharmacol Rev. 2013;65(1):315–499.
Pouton CW. Formulation of poorly water-soluble drugs for oral administration: physicochemical and physiological issues and the lipid formulation classification system. Eur J Pharm Sci. 2006;29(3–4):278–87.
Benson SP, Pleiss J. Molecular dynamics simulations of self-emulsifying drug-delivery systems (SEDDS): influence of excipients on droplet nanostructure and drug localization. Langmuir. 2014;30(28):8471–80.
Birru WA, Warren DB, Han S, Benameur H, Porter CJH, Pouton CW, Chalmers DK. Computational models of the gastrointestinal environment. 2. Phase behavior and drug Solubilization capacity of a type I lipid-based drug formulation after digestion. Mol Pharm. 2017;14(3):580–92.
Kasimova AO, Pavan GM, Danani A, Mondon K, Cristiani A, Scapozza L, Gurny R, Möller M. Validation of a novel molecular dynamics simulation approach for lipophilic drug incorporation into polymer micelles. J Phys Chem B. 2012;116(14):4338–45.
Larsson P, Alskär LC, Bergström CAS. Molecular structuring and phase transition of lipid-based formulations upon water dispersion: a coarse-grained molecular dynamics simulation approach. Mol Pharm. 2017;14(12):4145–53.
Marrink SJ, Tieleman DP. Molecular dynamics simulation of a lipid diamond cubic phase. J Am Chem Soc. 2001;123(49):12383–91.
Moghaddasi F, Housaindokht MR, Darroudi M, Bozorgmehr MR, Sadeghi A. Soybean oil-based nanoemulsion systems in absence and presence of curcumin: molecular dynamics simulation approach. J Mol Liq. 2018;264:242–52.
Rane SS, Anderson BD. Molecular dynamics simulations of functional group effects on solvation thermodynamics of model solutes in Decane and Tricaprylin. Mol Pharm. 2008;5(6):1023–36.
Suys EJA, Warren DB, Pham AC, Nowell CJ, Clulow AJ, Benameur H, Porter CJH, Pouton CW, Chalmers DK. A nonionic poly-ethyleneoxide (PEO) surfactant model: experimental and molecular dynamics studies of Kolliphor EL. J Pharm Sci. 2019;108(1):193–204.
Warren DB, Chalmers DK, Pouton CW. Structure and dynamics of glyceride lipid formulations, with propylene glycol and water. Mol Pharm. 2009;6(2):604–14.
Warren DB, King D, Benameur H, Pouton CW, Chalmers DK. Glyceride lipid formulations: molecular dynamics modeling of phase behavior during dispersion and molecular interactions between drugs and excipients. Pharm Res. 2013;30(12):1–16.
Warren DB, McPhee E, Birru WA, Benameur H, Chalmers DK, Pouton CW. Location of solvated probe molecules within nonionic surfactant molecules using molecular dynamics. J Pharm Sci. 2019;108(1):205–13.
Gao H, Jia H, Dong J, Yang X, Li H, Ouyang D. Integrated in silico formulation design of self-emulsifying drug delivery systems. Acta Pharm Sin B, 2021.
Holmboe M, Larsson P, Anwar J, Bergström CAS. Partitioning into colloidal structures of fasted state intestinal fluid studied by molecular dynamics simulations. Langmuir. 2016;32(48):12732–40.
Birru WA, Warren DB, Headey SJ, Benameur H, Porter CJH, Pouton CW, Chalmers DK. Computational models of the gastrointestinal environment. 1. The effect of digestion on the phase behavior of intestinal fluids. Mol Pharm. 2017;14(3):566–79.
Suys EJA, Warren DB, Porter CJH, Benameur H, Pouton CW, Chalmers DK. Computational models of the intestinal environment. 3. The impact of cholesterol content and pH on mixed micelle colloids. Mol Pharm. 2017;14(11):3684–97.
Hossain S, Kabedev A, Parrow A, Bergström CAS, Larsson P. Molecular simulation as a computational pharmaceutics tool to predict drug solubility, solubilization processes and partitioning. Eur J Pharm Biopharm. 2019;137:46–55.
Lüder K, Lindfors L, Westergren J, Nordholm S, Kjellander R. In silico prediction of drug solubility: 2. Free energy of solvation in pure melts. J Phys Chem B. 2007;111(7):1883–92.
Westergren J, Lindfors L, Höglund T, Lüder K, Nordholm S, Kjellander R. In silico prediction of drug solubility: 1. Free energy of hydration. J Phys Chem B. 2007;111(7):1872–82.
Brinkmann J, Luebbert C, Zaitsau DH, Verevkin SP, Sadowski G. Thermodynamic modeling of triglycerides using PC-SAFT. J Chem Eng Data. 2019;64(4):1446–53.
Brinkmann J, Exner L, Verevkin SP, Luebbert C, Sadowski G. PC-SAFT modeling of phase equilibria relevant for lipid-based drug delivery systems. J Chem Eng Data. 2021;66(3):1280–9.
Klamt A. Conductor-like screening model for real solvents: a new approach to the quantitative calculation of solvation phenomena. J Phys Chem. 1995;99(7):2224–35.
Paloncýová M, DeVane R, Murch B, Berka K, Otyepka M. Amphiphilic drug-like molecules accumulate in a membrane below the head group region. J Phys Chem B. 2014;118(4):1030–9.
Jakobtorweihen S, Zuniga AC, Ingram T, Gerlach T, Keil FJ, Smirnova I. Predicting solute partitioning in lipid bilayers: Free energies and partition coefficients from molecular dynamics simulations and COSMOmic. J Chem Phys. 2014. 141(4): p. 07B622_1.
Alsenz J, Kuentz M. From quantum chemistry to prediction of drug solubility in glycerides. Mol Pharm. 2019;16(11):4661–9.
Pozarska A, da Costa Mathews C, Wong M, Pencheva K. Application of COSMO-RS as an excipient ranking tool in early formulation development. Eur J Pharm Sci. 2013;49(4):505–11.
Bannigan P, Aldeghi M, Bao Z, Häse F, Aspuru-Guzik A, Allen C. Machine learning directed drug formulation development. Adv Drug Deliv Rev. 2021;175:113806.
Matsuda S, Koyasu S. Mechanisms of action of cyclosporine. Immunopharmacology. 2000;47(2):119–25.
Stuart MC, et al. Geneva: Geneva : world health organization. WHO model formulary. 2008:2009.
Dougados M, Awada H, Amor B. Cyclosporin in rheumatoid arthritis: a double blind, placebo controlled study in 52 patients. Ann Rheum Dis. 1988;47(2):127.
Brynskov JMD, et al. A placebo-controlled, double-blind, randomized trial of cyclosporine therapy in active chronic Crohn's disease. N Engl J Med. 1989;321(13):845–50.
Cattaneo D, Perico N, Remuzzi G. Generic cyclosporine formulations: more open questions than answers. Transpl Int. 2005;18(4):371–8.
McMillan MA. Clinical pharmacokinetics of cyclosporin. Pharmacol Ther. 1989;42(1):135–56.
Friman S, Bäckman L. A new microemulsion formulation of Cyclosporin. Clin Pharmacokinet. 1996;30(3):181–93.
Fahr A. Cyclosporin clinical pharmacokinetics. Clinical Pharmacokinetics. 1993;24(6):472–95.
Hauer B, Meinzer A, Posanski U, Vonderscher J. Cyclosporin compositions for oral administration. I.P. Office. 1992, Sandoz Ltd: United Kingdom.
Mueller EA, Kovarik JM, van Bree JB, Grevel J, Lücker PW, Kutz K. Influence of a fat-rich meal on the pharmacokinetics of a new Oral formulation of cyclosporine in a crossover comparison with the market formulation. Pharm Res. 1994;11(1):151–5.
Hauer B, Meinzer A, Posanski U, Richter F. Pharmaceutical compositions comprising cyclosporins, USPTO. 1995, Novartis Corporation: United States.
Choc MG. Bioavailability and pharmacokinetics of cyclosporine formulations: Neoral® vs Sandimmune®. Int J Dermatol. 1997;36(s1):1–6.
Kovarik JM, Mueller EA, Kutz K, van Bree JB, Tetzloff W. Reduced inter- and Intraindividual variability in cyclosporine pharmacokinetics from a microemulsion formulation. J Pharm Sci. 1994;83(3):444–6.
Williams HD, Sassene P, Kleberg K, Bakala-N'Goma JC, Calderone M, Jannin V, Igonin A, Partheil A, Marchaud D, Jule E, Vertommen J, Maio M, Blundell R, Benameur H, Carrière F, Müllertz A, Porter CJH, Pouton CW. Toward the establishment of standardized in vitro tests for lipid-based formulations, part 1: method parameterization and comparison of in vitro digestion profiles across a range of representative formulations. J Pharm Sci. 2012;101(9):3360–80.
Williams HD, Sassene P, Kleberg K, Calderone M, Igonin A, Jule E, Vertommen J, Blundell R, Benameur H, Müllertz A, Pouton CW, Porter CJH. Toward the establishment of standardized in vitro tests for lipid-based formulations, part 3: understanding supersaturation versus precipitation potential during the in vitro digestion of type I, II, IIIA, IIIB and IV lipid-based formulations. Pharm Res. 2013;30(12):3059–76.
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25.
Pall S, Abraham MJ, Kutzner C, Hess B, Lindahl E. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS, in Solving Software Challenges for Exascale, S. Markidis and E. Laure, Editors. 2015, Springer: Stockholm, Sweden p 3-27.
Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem. 2004;25(13):1656–76.
Warren DB, McPhee E, Birru WA, Benameur H, Chalmers DK, Pouton CW. Improvement in the predicted partitioning of alkane, alcohol and polyethylene oxide groups between water and octanol (logP) in molecular dynamics simulatoins. J Pharm Sci. 2019;108(1):214–22.
Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J, Pullman B. Interaction models for water in relation to protein hydration, in Intermolecular forces, B. Pullman, Editor. 1981, D. Reidel Publishing Company: Dordrecht. p. 331–342.
Feenstra KA, Hess B, Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J Comput Chem. 1999;20(8):786–98.
Miyamoto S, Kollman PA. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13:952–62.
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–72.
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–93.
Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101.
Nose S, Klein ML. Constant pressure molecular-dynamics for molecular-systems. Mol Phys. 1983;50(5):1055–76.
Parrinello M, Rahman A. Polymorphic transitions in single-crystals - a new molecular-dynamics method. J Appl Phys. 1981;52(12):7182–90.
van der Spoel D, van Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM, Hess B, Feenstra KA, Lindahl E, van Drunen R, Berendsen HJC. Gromacs User Manual version 3.1.1. 2002, Department of Biophysical Chemistry: Groningen p 278.
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–9.
Birru WA, Warren DB, Ibrahim A, Williams HD, Benameur H, Porter CJH, Chalmers DK, Pouton CW. Digestion of phospholipids after secretion of bile into the duodenum changes the phase behavior of bile components. Mol Pharm. 2014;11(8):2825–34.
Marques MRC, Loebenberg R, Almukainzi M. Simulated biological fluids with possible application in dissolution testing. Dissolution Technologies. 2011;18(3):15–28.
Rowe RC, Sheskey PJ, Owen SC, Handbook of pharmaceutical excipients. 5th ed. London: Pharmaceutical Press. 2006:xxi, 918 p.
Truong-Cong T, Millart E, Tran LTC, Amenitsch H, Frebourg G, Lesieur S, Faivre V. A scalable process to produce lipid-based compartmented Janus nanoparticles with pharmaceutically approved excipients. Nanoscale. 2018;10(8):3654–62.
Fatouros DG, Karpf DM, Nielsen FS, Mullertz A. Clinical studies with oral lipid based formulations of poorly soluble compounds. Ther Clin Risk Manag. 2007;3(4):591–604.
Klyashchitsky BA, Owen AJ. Drug delivery Systems for Cyclosporine: achievements and complications. J Drug Target. 1998;5(6):443–58.
Mains J, Tian R, Tian W, McNaughton A.The impact of the fasted and the fed state on BCS Class II cyclosporine formulation performance in vitro, in PharmSci 360. 2018, American Association of Pharmaceutical Scientists: Washinton, USA.
Škulj S, Vazdar M. Calculation of apparent pKa values of saturated fatty acids with different lengths in DOPC phospholipid bilayers. Phys Chem Chem Phys. 2019;21(19):10052–60.
ACKNOWLEDGMENTS AND DISCLOSURES
Financial support from Lonza - Pharma Biotech & Nutrition Sciences (formerly Capsugel) is gratefully acknowledged. This work was supported by the Multi-modal Australian ScienceS Imaging and Visualisation Environment (MASSIVE).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 2997 kb)
(MP4 20,799 kb)
(MP4 39,415 kb)
(MP4 31,784 kb)
(MP4 45,930 kb)
(MP4 65,551 kb)
(MP4 89,674 kb)
(MP4 25,603 kb)
(MP4 33,601 kb)
(MP4 35,566 kb)
(MP4 40,754 kb)
Animations of Final Frames
Animations of Sandimmune® and Neoral® in 90% water, 90% fasted buffer, 90% fed buffer conditions corresponding to Fig. 7, and digested formulations in 90% water unionized and ionized corresponding to Fig. 8 are included. The molecular representations are; cyclosporine A atoms = spheres (color cyan = carbon, red = oxygen, white = polar hydrogen), alkane chains = orange solvent accessible surface, surfactants = red licorice, glycerides = orange licorice, fatty acids = brown licorice, bile salts = green licorice, and phospholipid = yellow licorice. Solvents (water, ethanol and propylene glycol) have been omitted for clarity.
Rights and permissions
About this article
Cite this article
Warren, D.B., Haque, S., McInerney, M.P. et al. Molecular Dynamics Simulations and Experimental Results Provide Insight into Clinical Performance Differences between Sandimmune® and Neoral® Lipid-Based Formulations. Pharm Res 38, 1531–1547 (2021). https://doi.org/10.1007/s11095-021-03099-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-021-03099-5