Abstract
The origin of the genetic code is probably the central problem of the studies on the origin of life. The key question to answer is the molecular mechanism that allows the association of the amino acids with their triplet codons. We proposed that the codon-anticodon duplex located in the acceptor stem of primitive tRNAs would facilitate the chemical reactions required to synthesize cognate amino acids from simple amino acids (glycine, valine, and aspartic acid) linked to the 3′ acceptor end. In our view, various nucleotide-A-derived cofactors (with reactive chemical groups) may be attached to the codon-anticodon duplex, which allows group-transferring reactions from cofactors to simple amino acids, thereby producing the final amino acid. The nucleotide-A-derived cofactors could be incorporated into the RNA duplex (helix) by docking Adenosine (cofactor) into the minor groove via an interaction similar to the A-minor motif, forming a base triple between Adenosine and one complementary base pair of the duplex. Furthermore, we propose that this codon-anticodon duplex could initially catalyze a self-aminoacylation reaction with a simple amino acid. Therefore, the sequence of bases in the codon-anticodon duplex would determine the reactions that occurred during the formation of new amino acids for selective binding of nucleotide-A-derived cofactors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genetic code, the association between amino acids and trinucleotides (codons), shows that the codon arrangement in the standard genetic code table is non-random, i.e., related codons (those that differ at only one of the three positions) are assigned to similar amino acids.
Three major theories propose an explanation for the origin of the genetic code based on the nature of the driving force for amino acid assignments to the codons: the stereochemical theory (direct chemical interactions between RNA–codons and/or anticodons- and some amino acids) (Woese 1965; Shimizu 1982; Szathmary 1993; Yarus 1998; Yarus et al. 2005; Koonin and Novozhilov 2017), coevolution theory (biosynthetic pathways for generating new amino acids from early amino acids covalently attached to pre-tRNAs, leading to the development of the genetic code) (Wong 1975; Di Giulio 1994, 1998, 2008), and adaptive theory (adaptation-error minimization) (Freeland and Hurst 1998; Massey 2008; Higgs 2009). According to frozen accident theory the current codon assignment is mainly a historical accident and “yet related amino acids would be expected to have related codons” (Crick 1968).
Moreover, it has been suggested that these models are not mutually exclusive. Several models resulting from a combination of these basic models have been proposed (Knight et al. 1999; Copley et al. 2005; Kun and Radványi 2018). One of these models proposed that specific dinucleotides could have catalyzed the chemical reactions converting alpha-keto acids (covalently attached to dinucleotides) to specific amino acids, i.e., to their cognate amino acids (Copley et al. 2005), utilizing some strategies used by ribozymes.
It is generally accepted that current tRNAs arose during the RNA stage by ancestral duplication of an RNA hairpin structure (Bloch et al. 1985; Möller and Janssen 1990; Di Giulio 1992). Direct duplication of the precursor hairpin RNA molecule would have generated two regions (with two identical trinucleotide sequences); one could have evolved into the anticodon loop region and the other into the acceptor stem region, together forming the area that houses the main determinants of tRNA identity (Möller and Janssen 1990; Di Giulio 2004). The first substantial, if indirect, evidence supporting this notion was presented as possible presence of the codon-anticodon pairs in the acceptor stem of primitive tRNAs (Rodin et al. 1996). More recent and compelling evidence in support of a common ancestor for both codes (i.e., the RNA operational code embodied mostly in the acceptor stem and the genetic code per se embodied in the anticodon (Schimmel et al. 1993)) was also reported (Rodin et al. 2009).
It is possible that during ancestral stages of life, given the abundance of simple amino acids (such as glycine, valine, aspartate, and glutamate) originating from prebiotic processes, these amino acids could interact with small RNAs (such as the RNA minihelix -pre-tRNA-) by allowing or facilitating the synthesis of aminoacyl-RNA molecules. The presence of codon-anticodon pairs in the 3´ end of RNA minihelix may have allowed the binding of amino acids to these pre-tRNAs. The emergence of primitive tRNA (with anticodons and codon-anticodon pairs in the acceptor stem) and the formation of catalytic tRNA dimers (Martínez-Giménez and Tabares-Seisdedos 2002) may have facilitated this process.
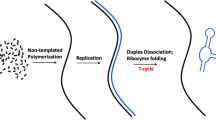
In this ancestral scenario, we propose a model for the origin of the genetic code comprising a similar A-minor motif interaction between cofactors derived from adenosine monophosphate (AMP) nucleotide (nucleotide-A-derived cofactor) and the complementary base pairs of the codon-anticodon duplex.This could determine the specific cofactors linked (united) to the catalytic tRNA dimer dependent on the base sequence in the codon-anticodon duplex (in the acceptor stem). Thus, this interaction (similar to the current A-minor motif) and the sequence of the codon-anticodon duplex could establish the order and nature of the cofactors attached to the codon-anticodon duplex, which would position the reactive groups (of the cofactors) close to the simple amino acid (attached by a covalent bond to the end of the acceptor stem in the tRNA dimer). Moreover, this would facilitate group-transferring reactions from cofactors to the simple amino acid by generating new amino acids. Thus, the nature of the cofactors attached to the codon-anticodon duplex (in the acceptor stem of the catalytic tRNA dimer), which would depend on the duplex sequence, could generate a new amino acids on the acceptor (see Fig. 1). This process would also depend on the simple amino acid initially attached to the tRNA dimer.
We propose that the codon-anticodon duplex of the tRNA catalytic dimer could facilitate the synthesis of new amino acids from simple amino acids using nucleotide-A-derived cofactors. RNAs can catalyze a wide range of chemical reactions (Cech et al. 1981; Zhang and Cech 1997; Jadhav and Yarus 2002; McGinness and Joyce 2003). Thus, the codon-anticodon duplex may catalyze reactions for the formation or modification of simple amino acids based on: (i) the positioning and electrostatics of reactants by hydrogen bonding interactions, (ii) the use of Mg2+coordinated to phosphate groups as catalysts, (iii) attachment of nucleotide-derived cofactors such as S-adenosylmethionine (SAM), flavin-adenine dinucleotide (FAD), coenzyme A (CoA) and phosphate of pyridoxal(PLP), (iv) the catalytic potential of the 2′-hydroxyl group in tRNA, (V) use of nucleobases to directly participate in general acid–base catalysis and (vi) the functional groups of modified bases in the anticodon. Many of these strategies are used by natural and selected ribozymes (Nakano et al. 2000; Adams et al. 2004; Das and Piccirilli 2005; Wolk et al. 2020; Minajigi and Francklyn 2008; Jiang et al. 2021).
In 1976, White proposed that current nucleotide-derived cofactors are relics of the catalytic center of ancestral ribozymes.
Recently, a small natural ribozyme that catalyzes the transfer of a methyl group from the SAM cofactor to a base was reported (Jiang et al. 2021). Furthermore, a natural pre-queuosine (Q1) riboswitch can catalyze the self-methylation reaction using m6preQ1 (O6-methyl-prequeuosine molecule) as a cofactor (Flemmich et al. 2021). These studies show that RNA can catalyze different reactions (unrelated to the phosphodiester bond), such as methylation, using small molecules as cofactors. One initial study showed that a natural cofactor-riboswitch has catalytic activity (Winkler et al. 2004). These findings support the proposal that some current riboswitches (that bind coenzymes) may be descendent from ancient ribozymes that synthesized these coenzymes or used to catalyze metabolic reactions (Breaker 2006; Cochrane and Strobel 2008).
Moreover, the FAD cofactor functions as a novel methylating agent in the methylation reaction catalyzed by the flavoprotein thymidylate synthase X FAD/folate-dependent (ThyX) enzyme (Hamdane et al. 2012; Mishanina et al. 2016).
A Cofactor-Based Mechanism for the Association of Amino Acids with their Codons
In this study, we propose a mechanism for the association between amino acids and their cognate codons. In our view, an initial scenario with pre-tRNAs (RNA minihelix) having codon-anticodon pairs in the 3´end and a high concentration of simple amino acids (such as glycine, valine, aspartate and glutamate) could allow the formation of specific aminoacyl-RNAs based on the weak interactions between glycine and a codon-anticodon pair of RNA minihelix. As noted above, primitive tRNA emergence (with anticodons and codon-anticodon pairs in the acceptor stem) and catalytic tRNA dimer formation (Martínez-Giménez and Tabares-Seisdedos 2002, 2021) could have facilitated this process.
Thus, we propose that glycine could have some preference for the codons of columns two and four of the code table, valine for codons in column one, and aspartate (glutamate) for codons in column three. Interestingly, one glycine-dependent riboswitch, only 86–126 nucleotides in size, has been reported (Mandal et al. 2004).
In this article, we propose that the codon-anticodon duplex located in the acceptor stem of primitive tRNAs facilitates the chemical reactions required to synthesize their cognate amino acids from simple amino acids (glycine, valine, glutamic acid and aspartic acid) linked to the 3′ acceptor end. In our view, various nucleotide-A-derived cofactors (with reactive chemical groups) may be attached to the codon-anticodon duplex, which allows group-transferring reactions from cofactors to simple amino acids, leading to the final amino acid (see Fig. 1). The nucleotide-A-derived cofactors could be incorporated into the RNA duplex (helix) by docking into the minor groove via an interaction similar to the A-minor motif, forming a base triple between A (cofactor) and one complementary base pair of the duplex (see Fig. 2). Tethering of the substrate (amino acid) and cofactor in close proximity via the codon-anticodon duplex might favor the transfer reaction. Furthermore, we propose that this codon-anticodon duplex could initially catalyze the self-aminoacylation reaction.
The self-aminoacylation process of tRNA with simple amino acids need not be specific, because some group-transferring reactions between the cofactors and this amino acid could not occur. Thus, valine binding to a tRNAgly molecule could not have an effect because the cofactor specific for this codon-anticodon duplex, pyridoxal phosphate (PLP), could not activate the carbon of valine.
The most likely group-transferring reaction is the addition of methyl groups to the initial amino acid, resulting in one new amino acid. Thus, a catalytic strategy could comprise the interaction of PLP (linked to the duplex by stacking between bases in the codon-anticodon helix) and the amino group of the amino acid forming a Schiff base, which could activate the (alpha)carbon (amino acid) and react with the methylene group of another FAD cofactor (attached to the base pair adjacent to the helix by A-minor interaction). Thus, a serine is produced from one initial glycine.
The sequence of bases in the codon-anticodon duplex would determine the types (nature) of reactions to form new amino acids for the selective binding of nucleotide-A-derived cofactors. Hence, the binding order of cofactors based on the triplet sequence would allow one specific reaction and the synthesis of a specific new amino acid (see Fig. 1).
Although the interaction of the A-minor motif is not very specific because the A-A dinucleotide in ribosomal RNA can recognize and interact with all correct types of Watson–Crick complementary base pairs in the short helix formed by interaction between tRNA anticodons and mRNA codons in the decoding process (during translation in ribosome) and in the tertiary structure of ribozymes (also the rRNA), a study showed that the interactions with C-G pairs predominated within ribosomal RNA (Nissen et al. 2001; Doherty et al. 2001). We propose that since the nucleotide-A-derived (5′-5′) cofactor is different from the A-A (3′-5′) dinucleotide of the decoding site (in ribosomal RNA), some cofactors may show a preference for the C-G base pairs of the codon-anticodon duplex and other cofactors for the A-U base pairs. In our view, the cofactors may have the following preferences in their interactions with the second base pair of codon-anticodon duplex: the FAD cofactor for G-C (Watson–Crick) base pairs (in the codon-anticodon duplex of tRNA dimer) of columns two and four of the code table (by A-minor interaction), the SAM cofactor for the A-U base pairs in the codons of column one (and some codons of three column; by stacking), and the CoA cofactor for the codons in columns three and four (by A-minor interaction). Other cofactors could have a low specificity, such as PLP and adenosylcobalamin (with a metal; AdoCbl). In short, these would be the basic rules that govern the interactions between the nucleotide-A-derived cofactors (adenosine) and the Watson–Crick base pairs.
Interestingly, in the natural ribozyme using SAM as a cofactor, the A base (of SAM) is inserted between A10 and U9 and stacked with A10 of the ribozyme (Jiang et al. 2021), i.e., these two bases are the closest to the A base of SAM. Other study (related with SAM) shows similar specificity for these A and U bases (Montange and Batey 2006).
Four types (variants) of the A-minor motif have been identified in the current RNAs (Nissen et al. 2001). In our vision, the FAD and CoA (and AdoCbl) nucleotide-A-derived cofactors could be incorporated into the RNA via an A-minor interaction of the type I between the adenosine (cofactor) and the second base pair of duplex (see Fig. 2). Moreover, we suggest that the SAM cofactor could have been incorporated in the RNA stacked between the first and second base pair of duplex. Thus, the second base pair could be regarded as a basic determinant in the nature of reactions occur via the selection of specific nucleotide-A-derived cofactor, in brief, a major, ancestral identity determinant of new formed amino acid. The present-day identity determinants could be the descendants of those ancestral determinants (in codon-anticodon duplex of pre-tRNA) that could have survived in the acceptor region of tRNAs.
These A-minor interactions are the most abundant tertiary structure interactions in the ribosomal RNA (Nissen et al. 2001). These interactions are basic in stabilizing the tertiary structure of RNA folds.
In summary, we propose that this primitive coding mechanism for new amino acid synthesis could be based on an interaction similar to the A-minor motif. This interaction is used in the current decoding mechanism of genetic information by the ribosome during genetic translation (Ogle et al. 2001).
Thus, we propose that the specificity of both amino acid–RNA interactions and cofactor–base pair (codon-anticodon duplex) interactions, although poor, could generate a reproducible pattern of reactions in the synthesis of new amino acids.
Although various studies have supported the prebiotic synthesis of some coenzymes or cofactors (Austin and Waddell 1999; Goldman and Kacar 2021; Kirschning 2021b), it has not yet been achieved.
One key fact is that the current biosynthesis of all 20 amino acids requires different sets of coenzymes, such as PLP, NAD+ (nicotinamide-adenine dinucleotide), CoA, and TPP (thiamine pyrophosphate). In the origin of life both molecules (amino acids and cofactors) could be formed independently of each other under prebiotic conditions (Kirschning 2021b). The prebiotic synthesis of at least several amino acids as the methionine occurs in variations of the classical Miller experiment (Bada 2013).
In one primitive scenario, coenzymes or cofactors could be partners of RNA rather than proteins (Kirschning 2021a, b). Moreover, the poor (catalytic) activity of pre-biotically formed coenzymes can be overcome by binding to short RNA fragments (Kirschning 2021a, b).
Furthermore, the notion that the coenzymes originated before the enzymes was suggested previously (Hartman 1975; White 1976). It has been suggested that ancestral cofactors (different from current cofactors) could function catalytically without proteins (along with metal ions and acting on surfaces supplied by clays) and with the capacity for self-replication, i.e., acting as the ancestral hereditary apparatus (Eakin 1963).
Another key element to establish a reproducible code is that each nucleotide-A-derived cofactor (which is universal) comprises an adenosine nucleoside and carries a different chemical structure with a chemically reactive group. Therefore, we propose that the reproducibility of binding of these two different chemical substances in the same molecule, the nucleotide-A-derived cofactor, is essential to establishing the genetic code. In our view, the presence of a conserved, universal group of nucleotide-A-derived cofactors was fundamental in the origin of a universal genetic code.
A recent study shows that NAD+ coenzymes are incorporated in bacterial RNA during the initiation of transcription by RNA polymerase (Bird et al. 2016). Other study showed that the NAD+ and 3´-desphospho-coenzyme A (dpCoA) coenzymes could only reside at the 5´end of RNA (Kowtoniuk et al. 2009). These works support the notion that the coenzymes could have been also involved in genetic (mechanism) processes in the origins of life.
Formation of some amino acids located in columns two and four of the genetic code table
Recent findings have shown that FAD can also mediate methyl transfer as a coenzyme of nucleic acid methyltransferases (Hamdane et al. 2012; Mishanina et al. 2016).
Based on this model, we propose that most codons in columns two and four of the code table initially bind glycine on the catalytic tRNA dimer (see Fig. 3). Moreover, the codon-anticodon pairs of the tRNA dimer could have simultaneously attached the FAD cofactor (on the second base pair of codon-anticodon duplex) and PLP (on the first base pair, closer to glycine). The A nucleoside of the FAD cofactor could have been incorporated at the codon-anticodon duplex (in the acceptor stem of tRNA) docked by an A-minor interaction, forming a base triple with the central G-C base pair, which would allow the positioning of reactive groups of the cofactor in proximity to glycine. The aldehyde group of PLP could form an imine (Schiff base) linkage with the amino group of glycine, which would activate one C-H bond, i.e., the deprotonation of the alpha carbon (glycine) with the formation of one carbanion stabilized by PLP. Subsequently, the alfa-carbanion of glycine (stabilized by PLP) could attack the methylene (-CH2OH) group of the FAD cofactor, yielding the serine amino acid (see Fig. 3).
Some steps of cofactor-based mechanism proposed for the formation of serine amino acid from the glycine by tRNA dimer. The structure of primitive tRNAs is proposed to have been similar to current tRNAs. a Binding of amino acid to the primitive tRNA. b Selective binding of two nucleotide-derived cofactors by codon-anticodon duplex. Activation of amino acid by PLP cofactor. c Group-transferring reaction from FAD(N) = CHOH cofactor to the activated amino acid and formation of new amino acid (serine)
We propose that all the codons of column two yield serine, except for the GCN (N = A, G, U, and C) codons, with a higher specificity for the FAD = CH2 cofactor generating alanine.
Moreover, we propose that the ACN codons (in column two) could undergo one additional (methyl) group-transferring reaction using two cofactors (FAD and other), yielding one threonine from the initial serine. In this case, the codon-anticodon duplex would have a specific affinity for these cofactors. The FAD = CH2 cofactor could interact with the second base pair of the codon-anticodon duplex (by an A-minor interaction). We propose that one cofactor (possibly with one metal ion in their molecule, similar to the adenosyl-cobalamin) would interact with the first base pair of codon-anticodon duplex (by similar an A-minor interaction), generating one activated beta-carbon (of serine) that could attack the methyl group in the FAD = CH2 cofactor, thereby producing threonine. One possibility is that only the ACN codons of the column showed an affinity for these cofactors.
In organic chemistry, a catalyst (with metal ions in the molecule) has been developed to convert the carbon-hydrogen (C-H) bonds of complex molecules into functional groups (with the addition of a methyl group) (Corcoran and Schultz 2020). Moreover, in some cases, organic catalysts resemble natural cofactors; thus, new catalysts have been developed from existing cofactors using a chemomimetic approach (Prier and Arnold 2015). Cofactors such as the current adenosyl-cobalamin but simpler than it could have played a role in threonine synthesis.
Moreover, we propose that most codons in column four would produce serine, except the GGN codons that generate glycine, which could bind the FAD and PLP cofactors, but the final structure would not have sufficient capacity to catalyze chemical reactions. Moreover, we propose that the tRNA dimers with UGY (Y = U and C; in column four) in the codon-anticodon duplex could undergo one additional reaction using acetyl-CoA as a cofactor by generating O-acetyl-serine, which could subsequently react with H2S to yield cysteine.
Formation of amino acids located in column one of the genetic code table
Based on this model, we propose that most column one codons would initially bind valine to the catalytic tRNA dimer. Moreover, the codon-anticodon pairs of tRNA dimers could have simultaneously attached the SAM cofactor (stacked between the first and second base pair of the codon-anticodon pair) and other cofactor similar to adenosyl-cobalamin (with one metal ion). This latter cofactor would have been incorporated at tRNA by an A-minor interaction with the first base pair of the codon-anticodon duplex. This would position the reactive (metal ion) groups of the cofactor in proximity to the valine, facilitating the activation of a C-H bond in the last carbon. We propose that valine’s activated carbon atom could attack the methyl group of the SAM cofactor to yield the isoleucine amino acid through one methyl-group transfer from the SAM cofactor to valine (see Fig. 4).
Potential cofactor-based mechanism for the formation of isoleucine amino acid from the valine by tRNA dimer. The structure of primitive tRNAs is proposed to have been similar to current tRNAs. a Binding of amino acid to the primitive tRNA. b Selective binding of two nucleotide-derived cofactors by codon-anticodon duplex. Activation of amino acid by cofactor similar to adenosyl-cobalamine. c Group-transferring reaction from SAM cofactor to the activated amino acid. d Formation of new amino acid (isoleucine)
Given the affinity of the SAM cofactor with the codons of the first column of the genetic code table, it possibly had a somewhat different interaction with the AUG codons of the codon-anticodon duplex without amino acid attachment. Therefore, on this occasion, the transfer of a methyl group would not occur; however, the complete methionine amino acid would be transferred from the SAM cofactor to the codon-anticodon duplex.
Formation of some amino acids located in column three of the genetic code table
In our view, we suggest that all codons of the column three would initially bind to aspartic acid and glutamic acid, as in the current GAN codons.
The AAR ( R = A or G) codon could bind to glutamic acid in the codon-anticodon duplex located at the 3′ end of the tRNA dimer. Moreover, the codon-anticodon pairs of the tRNA dimer could simultaneously attach to the acetyl-CoA cofactor (on the second or third base pair of the codon-anticodon duplex) and other cofactor (on the first or second base pair of the codon-anticodon duplex). The cofactor attached to the first or second base pair could facilitate the activation of one carbon and decarboxylation of glutamic acid. The modified bases of anticodons could also facilitate this process. We propose that the activated carbon could attack the reactive group of the acetyl-CoA cofactor, which would allow the acyl group (2 C) transfer to the amino acid residue. Subsequently, the NADH+ cofactor could have been incorporated at these codon-anticodon pairs to facilitate keto group reduction. Finally, other cofactors may have been involved in the addition of amino groups, which allowed the formation of lysine amino acids from glutamic acid.
Other type of cofactor could have been involved in adding one amino group to both aspartic acid and glutamic acid in their respective codons, generating asparagine (in AAC and AAU codons) and glutamine (in CAA and CAG codons).
Discussion
The RNA world hypothesis (Gilbert 1986) is currently the most accepted notion on the origin and evolution of the molecular genetic systems, in which RNA molecule functioned as both catalyst and (informational molecule) template. The model postulates one early era of self-replicanting RNAs which was later followed by the emergence of the genetic translation.Various models on the nature of the polymerase ribozyme have been proposed (Campbell 1991; Gordon 1995; Poole et al. 1998).
The RNA world notion is supported by the discovery of natural catalytic RNAs (Cech et al. 1981; Guerrier-Takada et al. 1983) and the finding that ribosomes are macromolecular machines with a RNA-based catalytic center (Ban et al. 2000).
The numerous discoveries of metabolite-sensing riboswitches and the studies that show that in vitro-selected and natural RNAs can catalyze reactions using small molecules as cofactors (including SAM cofactor) (Winkler et al. 2004; Scheitl et al. 2020; Jiang et al. 2021; Flemmich et al. 2021) support the notion that a complex biochemistry could have evolved in this RNA world. One exciting hypothesis is that present-day riboswitches for common cofactors may have evolved from ancient ribozymes that synthesized these coenzymes or used them to catalyze metabolic reactions (Breaker 2006; Cochrane and Strobel 2008). One key fact is that the current biosynthesis of all 20 amino acids requires different sets of coenzymes, such as PLP, NAD, CoA, and TPP. In the origin of life both molecules (amino acids and cofactors) could be formed independently of each other under prebiotic conditions (Kirschning 2021b). The prebiotic synthesis of at least several amino acids as the methionine occurs in variations of the classical Miller experiment (Bada 2013). Moreover, the FAD cofactor functions as a novel methylating agent in the methylation reaction catalyzed by the flavoproteinthymidylate synthase X FAD/folate-dependent (ThyX) enzyme (Hamdane et al. 2012; Mishanina et al. 2016). These data would support our model for the origin of genetic code. Moreover, a recent study shows that NAD+ coenzymes are incorporated in bacterial RNA during the initiation of transcription by RNA polymerase (Bird et al. 2016).
In our view one stage prior to the origin of genetic code was the emergence of nucleotide-derived cofactors, i.e., one stable community of molecules. Moreover, the bipartite structural feature of the nucleotide-A-derived cofactors was a basic element in the origin of the code.
In one RNA-based stage the RNA-catalyzed CoA, SAM, and FAD synthesis from their precursors (4´-phosphopantetheine, methionine, and FMN, respectively) it is possible. A study shows that RNAs are able to catalyze the synthesis of various coenzymes (CoA, FAD and NAD) (Huang and Yarus 2000).
Although the model above we have presented in relation to a scenario with fully formed tRNA, it may also be compatible with an earlier minihelix-based scenario (see Fig. 1).
In our view on the origin of code, the early association (matching) between amino acids and tRNA precursors could be driven by two types of direct interactions:
-
1.
The direct stereochemical interaction between the simple amino acids (glycine, valine and aspartic acid) and the codon-anticodon duplex in the end of the (pre-tRNA) minihelix. This minihelix could have functioned as a self-aminoacylating ribozyme. This interaction had low affinity and the high concentration of the simple amino acids could have favored it.
-
2.
One direct interaction between the adenosine of nucleotide-A-derived cofactor and the central base pair of codon-anticodon duplex (in the RNA minihelix with attached amino acid) via the A-minor motif of type I (with the formation of base triple) (see Fig. 2). In this interaction the adenosine N1, C2, N3 and 2´hydroxyl (O2´) groups interact (through hydrogen bonds) with the entire minor groove surface of second base pair (codon-anticodon duplex), including both riboses (Doherty et al. 2001; Nissen et al. 2001). The adenosine of SAM cofactor could have been incorporated at the end of RNA minihelix stacked between pair bases in duplex.
In contrast to other models on the origin of code, in our proposition the association (matching) of amino acids with their codons was conducted mainly in the language of nucleic acids via the A-minor interaction, one base triple between adenosine (cofactor) and Watson–Crick base pair (RNA). According with the model the information used in the synthesis of new amino acids is stored in the sequence of the pre-tRNA (minihelix) molecule and located in their 3´ end. This stored information in the sequences (pre-tRNA) located at acceptor end (codon-anticodon duplex) of molecule is read by the one-half of the cofactor (the A nucleotide) and executed by the other-half of the cofactor (the part with the reactive chemical group).
In our view genetic information stored in the RNA sequence was processed by nucleotide-A-derived cofactors. The capacity to process the information was possible by the bipartite structure of the nucleotide-A-derived cofactor. The nucleotide one-half of cofactor allows the recognition and binding of one specific base pair (in the codon-anticodon duplex). Immediately after this correct binding of cofactor to RNA minihelix, the reactive group-carrying other-half was positioned in such a way that the chemical group-transferring reaction from cofactor to amino acid can take place (can be carried out). The instructions for carrying out a particular chemical reaction and the instructions for reading the RNA sequence were embodied in the same molecule, the nucleotide-derived cofactor. In our model the role played by the nucleotide-A-derived cofactors is fundamental.
In the model, as in each of the chemical reactions in the synthesis of the amino acids two cofactors are involved simultaneously, it is possible that this operating mode could have determined the size of the codons as one triplet to be able to bind two cofactors each time. One nucleotide-A-derived cofactor (cofactor 1) should allow the activation of amino acid 1 and other nucleotide-A-derived cofactor (cofactor 2) would serve as carrier of a specific functional group (see Fig. 1). On our view, the coding mechanism was based in the selective binding of nucleotide-A-derived cofactors by the codon-anticodon duplex (Fig. 1). Moreover, the codon-anticodon duplex could bring together (in correct positioning) a group(reactive)-carrying cofactor (cofactor 2) and the activated amino acid 1 allowing the catalysis, i.e., the group-transferring reaction from cofactor 2 (auxiliary catalyst) to amino acid 1 generating a new amino acid (amino acid 2) (see Fig. 1). This process can be repeated two or more times during the synthesis of more complex amino acids as the lysine.
We posit that the capacity of the FAD cofactor to function as a methylating agent could be regarded as a molecular fossil of their role during the origin of genetic code.
Furthermore, the binding order of cofactors through the triplet sequence would allow one specific reaction and one synthesis specific to one new amino acid.
Other model point out also that the bases and their sequence are basic in the origin of genetic code (Copley et al. 2005, 2007).
The other regions of the self-aminoacylating ribozyme molecule could bind cofactors, but only the three base pairs at the 3′ end of the molecule would allow the attachment of the cofactors (with the adenosine nucleoside) close to the amino acid covalently bound to the 3′ end of the ribozyme, allowing specific chemical reactions on the amino acid.
The metabolic primitive pathways proposed for amino acid synthesis should have fewer steps and different routes than the current pathways. However, similar sets of coenzymes are required in both metabolic processes. In other words, only the use of nucleotide cofactors would have been preserved over time by nature.
It is possible to think that nature of current 20 natural amino acids was conditioned by the chemical nature of the initial nucleotide-A-derived cofactors.
In our model, the addition of methyl groups to different amino acids originating from prebiotic processes is a key element in the development of this genetic code. Although it is difficult to obtain many cofactors by synthesis under prebiotic conditions, there seem to be indications that this is possible.
The amino acids could have played a role important in the RNA world related with an increase of catalytic activity of RNAs (Gibson and Lamond 1990; Szathmary 1993; Roth and Breaker 1998; Grosjean et al. 2004).
References
Adams PS, Stahley MR, Wang J, Strobel SA (2004) Crystal structure of aself-splicing group I intron with both exons. Nature 430:45–50
Austin SM, Waddell TG (1999) Prebiotic synthesis of vitamin B6-type compounds. Orig Life EvolBiosph 29:287–296. https://doi.org/10.1023/a:1006532518221
Bada JL (2013) New insights into prebiotic chemistry from Stanley Miller´s spark discharge experiments. ChemSoc Rev 42:2186–2196
Ban N, Nissen P, Hasen J, Moore PB, Steiz TA (2000) The complete atomic structure of the large ribosomal subunit at 2,4A resolution. Science 289:905–920
Bird JG, Zhang Y, Tian Y, Panova N, Barvik I, Greene L et al (2016) The mechanism of RNA 5´capping with NAD+, NADH and desphospho-CoA. Nature 535:444–447
Bloch DP, Mcartur B, Mirrop S (1985) tRNA – rRNA sequence homologies: evidence from an ancient modular format by tRNAs and rRNAs. Biosystem 17:209–255
Breaker RR (2006) Riboswitches and the RNA world. In: The RNA world, 3rd ed., Gesteland RF, Cech TR, Atkins JE, eds., Cold Spring Habor Laboratory Press, pp.89–107
Campbell JH (1991) An RNA replisome as the ancestor of the ribosome. J MolEvol 32:3–5. https://doi.org/10.1007/BF02099922
Cech TR, Zaug AJ, Grabowski PJ (1981) In vitro splicing of the ribosomal RNA precursor of Tetrahymena:involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27:487–496. https://doi.org/10.1016/0092-8674(81)90390-1
Cochrane JC, Strobel SA (2008) Riboswitch effectors as protein enzyme cofactors. RNA 14:993–1002. https://doi.org/10.1261/rna.908408
Copley SD, Smith E, Morowitz HJ (2005) A mechanism for the association of amino acids with their codons and the origin of the genetic code. Proc Natl Acad Sci USA 102:4442–4447. https://doi.org/10.1073/pnas.0501049102
Copley SD, Smith E, Morowitz HJ (2007) The origin of the RNA world: co-evolution of genes and metabolism. Bioorg Chem 35:430–443
Corcoran EB, Schultz DM (2020) Methyl groups make a late entrance. Nature 580:592–593. https://doi.org/10.1038/d41586-020-01167-1
Crick FHC (1968) The origin of the genetic code. J MolBiol 38:367–379. https://doi.org/10.1016/0022-2836(68)90392-6
Das SR, Piccirilli JA (2005) General acid catalysis by the hepatitis delta virus ribozyme. Nature Chem Biol 1:45–52
Di Giulio M (1992) On the origin of the tRNA molecule. J Theor Biol 159:199–214
Di Giulio M (1994) On the origin of protein synthesis: a speculative model based on hairpin RNA sequences. J TheorBiol 171:303–308. https://doi.org/10.1006/jtbi.1994.1233
Di Giulio M (1998) Reflections on the origin of the genetic code: a hypothesis. JTheorBiol 191:191–196. https://doi.org/10.1006/jtbi.1997.0580
Di Giulio M (2004) The origin of the tRNA molecule: implications for the origin of protein synthesis. J TheorBiol 226:89–93. https://doi.org/10.1016/j.jtbi.2003.07.001
Di Giulio M (2008) An extension of the coevolution theory of the origin of the genetic code. Biol Direct 3:37. https://doi.org/10.1186/1745-6150-3-37
Doherty EA, Batey RT, Masquida B, Doudna JA (2001) A universal mode of helix packing in RNA. Nat StructBiol 8:339–343. https://doi.org/10.1038/86221
Eakin RE (1963) An approach to the evolution of metabolism. ProcNatlAcadSci USA 49:360–366. https://doi.org/10.1073/pnas.49.3.360
Flemmich L, Heel S, Moreno S, Breuker K, Micura R (2021) A natural riboswitch scaffold with self-methylation activity. Nat Commun 12:3877. https://doi.org/10.1038/s41467-021-24193-7
Freeland SJ, Hurst LD (1998) The genetic code is one in a million. J MolEvol 47:238–248
Gibson TJ, Lamond AI (1990) Metabolic complexity in the RNA world and implications for the origin of protein synthesis. J MolEvol 30:7–15. https://doi.org/10.1007/BF02102448
Gilbert W (1986) The RNA world. Nature 319:818
Goldman AD, Kacar B (2021) Cofactors are remnants of life’s origin and early evolution. JMolEvol 89:127–133. https://doi.org/10.1007/s00239-020-09988-4
Gordon KHL (1995) Were RNA replication and translation directly coupled in RNA (protein?) world? J Theor Biol 173:179–193
Grosjean H, de Crécy-Lagard V, Björk GR (2004) Aminoacylation of the anticodon stem by a tRNA-synthetaseparalog: relic of an ancient code? Trends BiochemSci 29:519–522. https://doi.org/10.1016/j.tibs.2004.08.005
Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–857
Hamdane D, Argentini M, Cornu D, Golinelli-Pimpaneau B, Fontecave M (2012) FAD/folate-dependent tRNAmethyltransferase: flavin as a new methyl-transfer agent. J Am ChemSoc 134:19739–19745. https://doi.org/10.1021/ja308145p
Hartman H (1975) Speculations on the origin and evolution of metabolism. J MolEvol 4:359–370. https://doi.org/10.1007/BF01732537
Higgs PG (2009) A four-column theory for the origin of the genetic code: tracing the evolutionary pathways that gave rise to an optimized code. Biol Direct 4:16
Huang F, Yarus M (2000) RNA-catalyzed CoA, NAD, and FAD synthesis from phosphopantetheine, NMN, and FMN. Biochemistry 39:15548–15555
Jadhav VR, Yarus M (2002) Acyl-CoAs from coenzyme ribozymes. Biochemistry 41:723–729. https://doi.org/10.1021/bi011803h
Jiang H, Gao Y, Zhang L, Chen D, Gan J, Murchie AIH (2021) The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences. Nat Catal 4:872–881. https://doi.org/10.1038/s41929-021-00685-z
Kirschning A (2021a) Coenzymes and their role in the evolution of life. AngewChemInt Ed Engl 60:6242–6269. https://doi.org/10.1002/anie.201914786
Kirschning A (2021b) The coenzyme/protein pair and the molecular evolution of life. Nat Prod Rep 38:993–1010. https://doi.org/10.1039/d0np00037j
Knight RD, Freeland SJ, Landweber LF (1999) Selection, history and chemistry: the three faces of the genetic code. Trends BiochemSci 24:241–247. https://doi.org/10.1016/s0968-0004(99)01392-4
Koonin EV, Novozhilov AS (2017) Origin and evolution of the universal genetic code. Annu Rev Genet 51:45–62. https://doi.org/10.1146/annurev-genet-120116-024713
Kowtoniuk WE, Shen Y, Heemstra JM, Liu DR (2009) A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc Natl Acadsci USA 106:7768–7773
Kun A, Radványi A (2018) The evolution of the genetic code: impasses and challenges. Biosystems 164:217–225
Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR (2004) A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306:275-279
Martínez Giménez JA, Tabares Seisdedos R (2002) On the dimerization of the primitive tRNAs: implications in the origin of genetic code. J Theor Biol 217:493-498
Martínez-Giménez JA, Tabares-Seisdedos R (2021) Possible ancestral functions of the genetic and RNA operational precodes and the origin of the genetic code. Orig Life EvolBiosph 51:167–183
Massey SE (2008) A neutral origin for error minimization in the genetic code. J MolEvol 67:510–516
McGinness KE, Joyce GF (2003) In search of an RNA replicase ribozyme. ChemBiol 10:5–14. https://doi.org/10.1016/s1074-5521(03)00003-6
Minajigi A, Francklyn CS (2008) RNA-assisted catalysis in a protein enzyme: The 2′-hydroxyl of tRNA–Thr A76 promotes aminoacylation by threonyl-tRNAsynthetase. ProcNatlAcadSci USA 105:17748–17753. https://doi.org/10.1073/pnas.0804247105
Mishanina TV, Yu L, Karunaratne K, Mondal D, Corcoran JM, Choi MA, Kohen A (2016) An unprecedented mechanismof nucleotide methylation in organisms containingthyX. Science 351:507–510. https://doi.org/10.1126/science.aad0300
Möller W, Janssen GMC (1990) Transfer RNAs for primordial amino acids contain remnants of a primitive code at position 3 to 5. Biochimie 72:361–368
Montange RK, Batey RT (2006) Structure of the S-adenosylmethionineriboswitch regulatory mRNA element. Nature 441:1172–1175
Nakano S, Chadalavada DM, Bevilacqua PC (2000) General acid–base catalysis in the mechanism of ahepatitis delta virus ribozyme. Science 287:1493–1497. https://doi.org/10.1126/science.287.5457.1493
Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA (2001) RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA 98:4899–4903. https://doi.org/10.1073/pnas.081082398
Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902
Poole AM, Jeffares DC, Penny D (1998) The path from the RNA world. J Mol Evol 46:1–17
Prier CK, Arnold FH (2015) Chemomimetic biocatalysis: exploiting the synthetic potential of cofactor-dependent enzymes to create new catalysts. J Am Chem Soc 137:13992–14006. https://doi.org/10.1021/jacs.5b09348
Rodin AS, Szathmáry E, RodinSN, (2009) One ancestor for two codes viewed from the perspective of two complementary modes of tRNAaminoacylation. Biol Direct 4:4. https://doi.org/10.1186/1745-6150-4-4
Rodin S, Rodin A, Ohno S (1996) The presence of codon-anticodon pairs in the acceptor stem of tRNAs. ProcNatlAcadSci USA 93:4537–4542. https://doi.org/10.1073/pnas.93.10.4537
Roth A, Breaker RR (1998)An amino acid as a cofactor for a catalytic polynucleotide. ProcNatlAcadSci USA 95:6027–6031. https://doi.org/10.1073/pnas.95.11.6027
Scheitl CP, Maghami MG, Lenz AK, Höbartner C (2020) Site-specific RNA methylation by a methyltransferase ribozyme. Nature 587:663–667
Schimmel P, Giegé R, Moras D, Yokoyama S (1993) An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci USA 90:8763–8768. https://doi.org/10.1073/pnas.90.19.8763
Shimizu M (1982) Molecular basis for the genetic code. J Mol Evol 18:297–303
Szathmary E (1993) Coding coenzyme handles; a hypothesis for the origin of the genetic code. Proc Natl Acad Sci USA 90:9916–9920
White HB (1976) Coenzymes as fossils of an earlier metabolic state. J MolEvol 7:101–104. https://doi.org/10.1007/BF01732468
Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR (2004) Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428:281–286. https://doi.org/10.1038/nature02362
Woese CR (1965) On the evolution of the genetic code. ProcNatlAcadSci USA 54:1546–1552. https://doi.org/10.1073/pnas.54.6.1546
Wolk SK, Mayfield WS, Gelinas AD, Astling D, Guillot J, Brody EN, Janjic N, Gold L (2020) Modified nucleotides may have enhanced early RNA catalysis. Proc Natl Acad Sci USA 117:8236–8242
Wong JT (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Yarus M (1998) Amino acids as RNA ligands: a direct-RNA -template theory for the code’s origin. J Mol Evol 47:109–117
Yarus M, Caporaso JG, Knight R (2005) Origins of the genetic code: the escaped triplet theory. Ann Rev Biochem 74:179–198
Zhang B, Cech TR (1997) Peptide bond formation by in vitro selected ribozymes. Nature 390:96–100. https://doi.org/10.1038/36375
Acknowledgements
Dedicated to my parents, Antonio and Maria Dolores (Juan). We thank a referee for comments on manuscript.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Martínez Giménez, J.A., Tabares Seisdedos, R. A Cofactor-Based Mechanism for the Origin of the Genetic Code. Orig Life Evol Biosph 52, 149–163 (2022). https://doi.org/10.1007/s11084-022-09628-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-022-09628-5