Abstract
Among various scenarios that attempt to explain how life arose, the RNA world is currently the most widely accepted scientific hypothesis among biologists. However, the RNA world is logistically implausible and doesn’t explain how translation arose and DNA became incorporated into living systems. Here I propose an alternative hypothesis for life’s origin based on cooperation between simple nucleic acids, peptides and lipids. Organic matter that accumulated on the prebiotic Earth segregated into phases in the ocean based on density and solubility. Synthesis of complex organic monomers and polymerization reactions occurred within a surface hydrophilic layer and at its aqueous and atmospheric interfaces. Replication of nucleic acids and translation of peptides began at the emulsified interface between hydrophobic and aqueous layers. At the core of the protobiont was a family of short nucleic acids bearing arginine’s codon and anticodon that added this amino acid to pre-formed peptides. In turn, the survival and replication of nucleic acid was aided by the peptides. The arginine-enriched peptides served to sequester and transfer phosphate bond energy and acted as cohesive agents, aggregating nucleic acids and keeping them at the interface.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent years have seen a renaissance in the study of life’s origins from both theoretical and experimental perspectives. Much excitement has surrounded the field of astrobiology, as explorations of Earth’s planetary neighbors in the solar system expand and our knowledge of other planetary systems increases (National Research Council 2007). After it was concluded that classic “prebiotic soup” scenarios seem unlikely to permit the formation of complex organic monomers and functional biopolymers on Earth, new chemical evolution models have been proposed and explored that address how prebiotic organic synthesis and polymerization may have occurred. These models include the “lipid world” (Cavalier-Smith 1987; Segré et al 2001) and possible roles of aerosols (Tervahattu et al. 2004), mineral surfaces (Cairns-Smith 1982; Deamer et al. 2006; Ferris 2006) and hydrothermal vents (Wächtershauser 2006). Scenarios that address the complicated questions of how biological information and the biological functions of metabolism, replication and translation arose have also seen much attention, two alternative scenarios currently being prominent. In “metabolism first” scenarios (DeDuve 1991; Wächtershauser 1992; Morowitz et al. 2000; Shapiro 2006) a complex web of organic chemical reactions involving small molecules preceded and eventually led to nucleic acid synthesis, replication and translation. Since “metabolism first” scenarios do not directly address how replication and translation arose, most biologists favor “replication first” models for the origin of biological function and life, specifically the “RNA world” scenario (Gilbert 1986; Line 2002).
The RNA world scenario proposes that RNA was the sole information-rich macromolecule, undergoing replication and catalyzing metabolic reactions prior to the occurrence of DNA and proteins. The RNA world was independently proposed by Woese (1967), Crick (1968) and Orgel (1968) to avoid the “chicken or egg” paradox of the primacy of metabolism (protein first) or replication (DNA first). Since the RNA world involves a single kind of information-rich molecule that is capable of both replication and catalysis it is viewed as more parsimonious than scenarios involving both nucleic acid (NA) and proteins. The RNA world received support from the discovery of catalytic ribozymes (Cech et al. 1981). It has been reinforced as additional catalytic roles of natural RNA were discovered (Doudna and Cech 2002) and laboratory experiments demonstrated RNA can evolve enhanced catalytic function under in vitro selection (Joyce 2002).
Despite its popularity and value in stimulating experimental studies, the RNA world has serious weaknesses, both logistical and conceptual. Simulations of prebiotic chemistry do not easily generate some RNA components or allow their polymerization, particularly if competing reactive substances are present (Miller 1987; Joyce and Orgel 1999; Shapiro 1999). The number and complexity of ribozymes deemed necessary for a minimal living system are logistically implausible (Benner et al. 1993; Joyce and Orgel 1999; Shapiro 1999) to the point where a proponent, perhaps with deliberate intent to be provocative, invoked the eternal inflation cosmological model to account for the RNA world (Koonin 2007). In addition to logistical issues, the RNA world does not directly explain the transition to the current DNA/RNA/protein world (Cavalier-Smith 1987). Various proposals have tried to account for the genetic code’s origin in a RNA world (e.g. Weiner and Maizels 1987; Yarus et al. 2005; Wolf and Koonin 2007; Yakhnin 2007) but no compelling explanation has yet arisen.
Because the RNA world is logistically implausible and has not led to convincing explanations for the genetic code’s origin or DNA’s incorporation into the genome, alternative scientific hypotheses for the origin of biological function should be considered. Several authors have supported the general idea that simple peptides and NAs could have arisen together and cooperatively led to the evolution of living systems (Kunin 2000; Nashimoto 2002), However, no specific scenario has yet been proposed addressing in detail how this might have occurred.
Here I develop a comprehensive and specific scenario for the origin of life based on the premise that simple peptides and nucleic acids arose simultaneously and life began through their interdependent activity. Since the physical and chemical environment was responsible for the generation, accumulation and interaction of component molecules of the first living system, both pre-biotic and proto-biotic environmental conditions are addressed. In developing the scenario I constructed a model of the protobiont, a simple hypothetical system that most would agree was “alive” and that suggests pathways through which complex organisms living today could evolve. Modeling a simple ancestor has proven useful in focusing on the issues surrounding other major evolutionary transitions with poor fossil evidence like the origin of vertebrates (Griffith 1994). The application of this technique to life’s origin has been advocated (Rosen 1984; Lahav 1993) and attempted (Jeffares et al. 1998), the RNA world being such a model.
Given a wide range of views and approaches to the origin of life among scientists, it is unlikely a consensus could be reached on minimal requirements for living systems. Since the model should explain how life as we know it arose on Earth, rather than define what generic properties life might have anywhere, I support the rigorous criteria listed below. Our model protobiont should (a) have a defined molecular composition, (b) replicate, (c) convert environmental energy into macromolecular bond energy, (d) be “cohesive”, allowing functional components to interact, (e) suggest how the genetic code originated, (f) be plausible in terms of realistic prebiotic conditions, (g) not rely on contrivances absent from extant living organisms, (h) be subject to evolution via natural selection, and (i) be testable, leading to experimental studies.
I will describe my scenario in four sections. I will outline the environmental context of life’s origins, describe general attributes of the protobiont, propose a specific model amenable to testing and finally suggest pathways of subsequent evolution towards organisms possessing complex metabolism and the full genetic code. The specific model will include the size and base sequence of NAs and the amino acid composition of peptides. In addition to structure, the scenario will explain how the model protobiont interacts with the environment and how it functions during replication, translation and metabolic energy transfer.
The Habitat of the Protobiont
In classic models of prebiotic chemical evolution, organic compounds formed in a reducing atmosphere with the aid of energy sources such as lightning (Urey 1952). These compounds accumulated in the sea to form an “organic soup”, allowing polymerization to biologically active molecules, including peptides and NAs. These subsequently coalesced into living systems.
In recent years the classic “organic soup” scenario is generally viewed as implausible. The presence of a sufficiently reducing atmosphere has been questioned (Miller 1987; Bada 2004), although recent reports suggest a reducing environment could have been present (Tian et al. 2005). Key monomers like sugars and some nucleobases are difficult to form under what are regarded as realistic prebiotic conditions (Orgel and Lohrman 1974; Shapiro 1999). Polymerization reactions that could lead to peptides or NAs do not occur in dilute aqueous media, requiring special concentrating and condensing mechanisms (Bada 2004). The possibility of getting functional information-rich molecules through stochastic processes is viewed as remote (Cairns-Smith 1982; Wächtershauser 2000; Koonin 2007). Furthermore, the conceptual questions of how biological functions of metabolism, inheritance and translation arose are equally or even more difficult than the logistical issues. The literature is replete with disparate attempts to explain how these logistical and informational problems could be avoided (Bada 2004), but collectively they would seem to be a serious challenge to the hypothesis that life arose on Earth through chemical evolution.
I contend there is a plausible and largely overlooked solution to most logistical and informational problems. Partitioning of organic substances at and near the prebiotic ocean’s surface could have led to the formation, selection and functional interaction of peptides, NAs and other molecules important to life.
Recent reviews have addressed the physical and chemical conditions on the prebiotic Earth (Bada 2004; Bernstein 2006; Lunine 2006). During Earth’s early history, an initial period of accretion was followed by a relatively quiescent period, and later the ‘late heavy bombardment’ that lasted until around 3.8 billion years ago. The latter may have vaporized the oceans, perhaps precluding survival of living organisms or complex organic compounds (Nisbet and Sleep 2001; Bada 2004). Our scenario commences soon after the last sterilizing impact; we assume a hot and gradually cooling oceanic planet (Sleep et al. 1989). A variety of processes might contribute to the pool of organic material present on Earth’s surface at the time including volcanism, hydrothermal activity, impact-related Fisher-Tropsch type syntheses, and atmospheric reactions driven by lightning, coronal discharge, ultraviolet radiation and impact shocks; extraterrestrial sources supplementing planetary synthesis (Prinn and Fegley 1989; Mason 1991; Chyba and Sagan 1992; Bada 2004; Bernstein 2006). The quantity and diversity of organic matter present would depend upon Earth’s atmospheric composition, the relative contribution of different synthetic processes and relative rates of synthesis and degradation—issues that are moot (Bernstein 2006). A reasonable working hypothesis is that the organic mixture on the Earth’s surface included the full inventory of substances that are produced in prebiotic simulation experiments and those occurring in carbonaceous meteorites (i.e. organic water-soluble substances including simpler amino and carboxylic acids, non-polar hydrocarbons and heterocyclic compounds and an insoluble kerogen of complex composition [Mason 1991; Bada 2004]). Furthermore, given the presence of life on Earth, it is reasonable to simply suggest that the diversity and quantity of organic substances were sufficient to allow life to arise somewhere on the planet.
As our scenario begins, most of Earth’s surface would have been covered with an ocean thousands of meters deep; the presence of continental crust at the time is questionable and terrestrial and shallow water habitats were likely of negligible global significance (Mason 1991; Lunine 2006). The ocean probably possessed higher salinity than at present (Knauth 1998) and was enriched with organic material. The latter partitioned into a superficial oil slick composed of hydrocarbons, esters and other low-density non-polar substances (Lasaga et al. 1971; Nilson 2002; Bada 2004), an aqueous layer with modest dissolved organic solute concentrations, and sediment of dense kerogen and organic precipitates. The boundary zone between the oil slick and the polar aqueous layer contained amphiphiles having both polar and non-polar domains (Nilson 2002).
The thickness of the oil slick, concentration of soluble organic molecules in the aqueous phase, amount of kerogen and quantity of amphiphiles are important but largely conjectural issues. Lasaga et al (1971) suggested the oil slick may have been 1–10 m thick, common carboxylic and amino acids in the ocean were probably in micromolar concentrations or less (Stribling and Miller 1987) and much of the total organic carbon likely was kerogen (Mason 1991). Amphiphiles may have constituted a substantial fraction of the total organic material (Deamer 1997; Segré et al. 2001). Organic matter would have undergone exchanges between phases. For instance, hydrolysis and pyrolysis would release oils and soluble monomers and polymers from kerogen, and a complex set of reactions within the oil slick and at its atmospheric and aqueous interfaces (discussed in more detail below) would generate polymers, hydrophilic molecules, amphiphiles and kerogen. Extraterrestrial input and atmospheric and hydrothermal synthesis of monomers would continue. The presence of abundant organic material and high salinity would have affected availability of inorganic ions of biological importance. For instance, binding of calcium by polyanionic components of kerogen or oxalic acid (Day 1984) as well as the inhibition of calcium phosphate mineralization at high salinities (Nancollas and Tomazik 1974; Hagen et al. 2007) would make phosphorus available for formation of organic phosphate compounds including NAs. Organo-metallic complexes containing iron or other ions could have been important components of the oil slick or amphiphiles, participating in prebiotic redox reactions.
Although some models of the early Earth’s heat budget favor an ice-covered planet (Bada 2004), the time frame of our “hot Earth” scenario and presence of an oil slick that would absorb solar energy argue against this. Absorption of solar energy would induce diurnal thermal cycles with temperatures near the surface of the oil slick exceeding the boiling point of water during the day but cooling at depth and by night. This energy absorption would also help generate reactive compounds (Nilson 2002). Aqueous droplets derived from both the atmosphere as rain and through convective processes at the oil/water interface would form “reverse micelles” within the oil slick, stabilized by amphiphilic molecules. Polar molecules within these reverse micelles (including amino acids, urea, formaldehyde and other solutes) would be subject to condensation and dehydration reactions driven by evaporation, thermal cycles and solar energy (Nilson 2002). Collectively, reactions within the oil slick and at its interfaces with the atmosphere and the aqueous phase could have led to the formation of a heterogeneous mixture of sugars, added to the pool of heterocyclic compounds, (including purines and pyrimidines) and formed a variety of amphiphiles including peptides, esters, nucleosides and organic phosphate compounds. Given the coincidence of sugars, nucleobases, and organic phosphate amphiphiles, as well as dehydrating conditions and solar energy, it is certainly reasonable that nucleic acids would be generated within the oil slick.
The boundary zone between oil slick and aqueous phase would be an emulsion rather than a clean interface, provided there were sufficient amphiphiles and convective forces near the ocean’s surface. In addition to reverse micelles within the oil slick, emulsified droplets would include micelles and vesicles within the aqueous zone (Segré et al. 2001). Convective forces would aid emulsification and also transport droplets vertically and horizontally, permitting fusion of droplets and exposing them to environmental gradients. Differential heating, changes in droplet density due to gain or loss of oils, and internal waves at the oil slick/aqueous interface would be among phenomena driving convective transport.
I favor the emulsified boundary zone between oceanic hydrophobic and aqueous layers as the site where life arose. The oil slick would not be a suitable place for bio-molecular functions like replication, translation and metabolic catalysis because these require hydrogen bonding or electrostatic attraction. Furthermore, functional macromolecules would be subject to ultraviolet damage were they within the oil slick. Biological function could not develop in the early ocean’s polar layer because of low organic concentrations and hydrolysis of polymers. The benthic kerogen layer, mineral surfaces (Cairns-Smith 1982; Wächtershauser 1988) and submarine hydrothermal vents (Shock 1992) offer surface area and could serve as sites for molecular accumulation and chemical reactions of potential biological significance, but they offer no obvious way to transition into living systems as we know them.
The emulsified oil/aqueous interface offers numerous features conducive to the origin of a familiar life. Emulsification droplets within the aqueous phase (small micelles, larger chylomicron-like particles and cell-like bodies formed by the enclosure of polar sea water inside a lipid ‘membrane’ stabilized by amphiphiles) offer enormous surface area and versatile structural stages for molecular accumulation and interaction (Deamer 1997; Segré et al. 2001). The oil slick generates activated ‘high energy’ compounds, complex building blocks and polymers, including NAs and peptides, and also provides protection of bio-molecules from degradation by high-energy solar radiation. The aqueous layer provides soluble organic units useful in building functional polymers and ions useful in metabolism. Internal waves that form between fluid layers of different densities, familiar to physical oceanographers and limnologists and implicated in aggregation and transport of plankton (Shanks 1983), could facilitate vertical and horizontal exchange, favor particulate and macromolecular aggregation and entrain cyclical molecular phenomena. The latter could include NA replication. Furthermore, amphiphilic molecules accumulating at the boundary zone would include peptides and organophosphate compounds compatible with the chemistry of extant living systems and able to maintain NAs at the site.
Perhaps one of the most compelling reasons for favoring the oceanic oil/aqueous interface for the origin and early evolution of life is that it is global in scope and allows rapid convective exchange, even over great distances. The origin of an interacting set of functional bio-molecules and the elaboration of the genetic code would seem to entail an improbable sequence of events of increasing complexity that I believe can only be explained by a grand experiment on a global scale, not within the constraints of Darwin’s “warm little pond” or comparably restricted locations.
General Features of the Protobiont I: Molecular Composition
The premise of my scenario is that life arose through interdependent activity of NA and peptides. I propose that the NA was a conventional NA with sugar/phosphate backbone and purine and pyrimidine bases able to form Watson/Crick hydrogen bonds. Some authors, working from the assumption that logistical difficulties preclude its prebiotic synthesis, have proposed that simpler versions (“TNA” or “PNA”) preceded conventional NAs (Nelson et al. 2000; Eschenmoser 2004). I believe such models are neither necessary nor parsimonious. Our reconstruction of the prebiotic Earth accounts for the generation of conventional NA. Scenarios using alternatives not only have to explain how these non-biological analogs arose and functioned, but also how and why they transitioned to systems based on conventional NA. For similar reasons we argue the first peptides were based on amino acids now used in protein.
Arguments favoring RNA as the original NA, implicit in the RNA world, include its ability to act as a catalyst and interact with protein, its primacy in modern biosynthetic pathways and its use as a primer during DNA replication (Maizels and Weiner 1987; Lazcano et al. 1992). I contend the question of which was first, RNA or DNA, is a red herring; both could be present and functional from life’s inception. Prebiotic syntheses producing ribose would also yield deoxyribose (and many other sugars) and, depending upon concentrations and conditions, conversions between sugars would occur. Thymine (as well as other nitrogenous bases not found in DNA and RNA today) would be synthesized under conditions that could generate uracil. The yield of pure RNA or DNA strands through non-template-based processes would be extremely low, only short oligomers being plausible. Longer strands would likely be heterogeneous in both sugar and kind of base and most reactions involving NA components would lead to ‘heteromers’ incorporating units other than nucleotides.
Thermal cycling in the emulsified boundary zone could promote template-based NA replication through Watson/Crick hydrogen bonding (discussed in more detail in the next section). This could lead to a pre-biological/pre-Darwinian selection for NA that formed stable double strands. Stereospecificity would favor the accumulation of true, homochiral NA relative to heterogeneous strands during template-based replication. Given high temperatures during the early stages of molecular evolution, DNA would accumulate relative to RNA because it is able to form protected double strands at higher temperatures and, lacking 2′ hydroxyl groups, is less subject to hydrolysis. High temperature would favor a preponderance of G-C pairs in double-stranded DNA. As temperatures cooled, RNA strands could be transcribed from DNA. In the absence of enzymes able to distinguish between DNA and RNA and their nucleotides, NAs that formed on templates would tend to be heterogeneous for the sugars ribose and deoxyribose, the composition depending on temperature and availability.
The presence of both DNA and RNA (plus heterogeneous strands) in the model provides a versatile starting point for molecular evolution. DNA provides a genome that is stable and able to replicate reliably, even at high temperatures. The involvement of RNA is favored because its turnover would be more rapid and its exposed bases and three dimensional shape allow it to interact with amino acids and peptides. With both DNA and RNA present from the beginning, the difficult question of how transcription originated is avoided.
Peptides enriched with basic amino acids, specifically arginine, are at the core of my model protobiont. The central premise of the scenario is that life originated as a set of interactions between NAs and peptides that was mutually beneficial—natural selection favoring the proliferation of NA families that helped incorporate specific amino acids into peptides which, in turn, facilitated NA survival and copying. DNA and RNA are water-soluble and do not self-aggregate, so that dispersal into the ocean’s aqueous phase represents a serious problem for a NA-based protobiont. Thus, mechanisms leading to NA aggregation and maintenance at the oil/water interface are essential. Electrostatic interactions of basic ‘R’ groups in peptides (i.e. the amine group of lysine, guanadinium group of arginine or imidazole group of histidine) with the phosphate backbones of NA could serve to aggregate single or double-stranded NA in the protobiont as ribosomal proteins and histones do in organisms today. Base-enriched peptides could also keep NAs at the emulsified zone between lipid and aqueous layers. Experimental data suggest basic proteins aid incorporation of NAs in lipid vesicles (Jay and Gilbert 1987). Base-enriched peptides would be most effective in this role if they possessed a substantial hydrophobic domain and multiple basic amino acid residues.
Of the three basic amino acids that now occur in protein, I favor the primacy of arginine. In our model both template-based NA replication and peptide bond formation use “high energy” phosphate transfer of the sort now centered on ATP metabolism. Histidine and lysine are able to bind phosphate electrostatically, but only arginine’s guanidinium group easily forms “high energy” phosphate covalent bonds. Mollusks and arthropods use phosphoarginine rather than phosphocreatine as their muscle phosphagen (Hochachka and Somero 1984). In our model, arginine-enriched peptides capture phosphate from reactive compounds generated through photochemistry in the oil slick and transfer it during NA copying or peptide bond formation. Arginine residues are crucial to the active sites of enzymes participating in diverse phosphate transfer reactions including pyruvate kinase (Muirhead 1987), creatine kinase (Wood et al. 1998) and aminoacylation reactions catalysed by tRNA synthetases (Arnez et al. 1997). Arginine “fingers” participate in nucleotide triphosphate hydrolysis through stabilization of the transition state of the reaction (Rittenger et al. 1997; Crampton et al. 2004).
Our suggestion that arginine was the first amino acid specified in translation is contrary to most scenarios for the origin of the genetic code which generally favor the primacy of simple amino acids produced abundantly in prebiotic syntheses (Trifonov 2004; Copley et al. 2005). Certainly arginine does not fit these descriptions. However, our scenario is based upon indispensable functional roles of the first translated amino acid in peptide/NA interactions and metabolism. We believe facile synthesis is irrelevant; amino acids as complex as arginine will readily be formed by the same processes that yield nucleobases—e.g. the thermal anhydrous condensation of molecules with short carbon skeletons (including urea) within reverse micelles.
General Features of the Protobiont II: Functional Biology
Although the following discussion focuses mostly on functional interactions between arginine-enriched peptides and one particular NA family, the importance of other organic molecules to our scenario must be reiterated. Lipids and amphiphiles at the emulsified zone favor accumulation of NA, peptides and building blocks; photoreactive molecules within the oil slick are energetically important; various small molecules serve as building blocks, substrates or intermediates in metabolism; non-functional mixed “NA” polymers act as nuclei for aggregation and are reservoirs of building blocks and bond energy; NA/peptide interactions involving amino acids other than arginine become components of the genome as the protobiont evolves. From this perspective the model protobiont includes all the useful molecules of the environment as well as the arginine translator, being essentially a global superorganism.
In our scenario, NA copying is initially driven by environmental thermal cycles with the support of base-enriched peptides. Depending upon pressure, solute concentrations and peptide interactions, cyclical fluctuation in temperature around the boiling point of water could induce NA melting, followed by renaturation, hybridization or template-based replication like that seen in the polymerase chain reaction. Diurnal cycles could lead to thermal variation on a hot early Earth, but molecular evolution would be slow if this were the only cause. Internal waves at the oil/water interface could provide thermal cycling of a suitably short period—seconds or minutes. Depending on amplitudes of internal waves and the thickness of the oil slick, emulsified droplets (either reverse micelles in the oil slick or micelles or vesicles in the aqueous layer) could undergo vertical oscillations subjecting them to considerable temperature variation, especially during daytime. Cyclical environmental fluctuations were previously suggested to drive non-enzymatic NA replication (Usher 1977; Lahav 1991). Internal waves at the aqueous-lipid interface represent a plausible specific mechanism of a suitable time scale.
Base-enriched peptides play several important roles in this process. They serve to protect NA from thermal degradation as do base-enriched histones in hyperthermophilic Archaea (Reeve et al. 2004). By virtue of their binding to the backbones of NA, base-enriched peptides could facilitate NA copying by aligning template and copy strands. Some experimental evidence suggests that simple base-enriched peptides can facilitate non-template-based elongation of NAs from activated monomers (Woese 1968; Barbier et al. 1993) raising the possibility of reverse translation. Furthermore, as described below, arginine residues could provide bond energy for the formation of NA phosphodiester bonds.
In contemporary organisms, macromolecular syntheses like replication, transcription and peptide synthesis use ATP and other triphosphonucleotides as bond energy sources. Because of high solubility and low stability in water, dissolved ATP could not have served as an environmental energy source. Membrane-based phosphorylation mechanisms that use proton gradients and electron transfer are implausible this early. The most reasonable bond energy sources for our model protobiont are polynucleotide fragments already possessing phosphodiester bonds and ‘high energy’ organic phosphate amphiphiles formed through photochemistry in the oil slick.
Replication in our model is achieved by aligning partially complementary NA fragments on template strands, linking these fragments through energetically neutral transesterification and excising non-complementary loose ends. Experimental studies suggest this could be an efficient copying mechanism for oligonucleotide DNA templates under the conditions of our scenario (James and Ellington 1997). Clearly, this process allows NA modifications like elongation as well as replication. If a full complement of fragments were unavailable, gaps could be bridged using individual nucleosides or nucleotides. Energy for new phosphodiester bonds is provided by phosphate transfer from peptidyl arginine residues. These, in turn, would be phosphorylated through transfer from high-energy phosphate amphiphiles generated photochemically in the oil slick. As discussed below, high-energy amphiphilic phosphates also provide energy for peptide bond formation.
In extant organisms translation requires numerous specialized macromolecules. Peptide elongation involves recognition and activation of 20 different amino acids by aminoacyl tRNA synthetases, acylation with appropriate tRNAs, alignment of amino acids relative to the growing peptide through tRNA/mRNA hydrogen bonding, and formation of peptide bonds linking amino acids to the peptide. Furthermore, translation must be initiated and it requires a structural framework and catalysis provided by ribosomes. Translation becomes very complicated if more than one kind of amino acid is coded and more than one kind of protein translated. In our simple model translation incorporates only one kind of amino acid (arginine) into one kind of peptide, uses no adaptor enzymes and involves complementary oligonucleotide strands that serve as tRNA and mRNA.
Recognition of arginine in our model protobiont involves direct NA/amino acid stereospecific binding without adaptor enzymes. Experimental data suggest recognition through direct interaction between amino acid and NA is quite feasible in the case of arginine; RNA aptamers show binding affinity in the sub-micromolar range and are highly eniatioselective (Geiger et al. 1996; Yarus et al. 2005). Sites within RNA aptamers responsible for high arginine affinity are correlated with this amino acid’s codons (Knight et al. 2003). Thus, arginine is recognized by a NA strand that displays at least one of its codons, ideally both motifs.
In our model, alignment of free arginine relative to an extant arginine-enriched peptide involves two complementary NA strands. The peptide and NA are held together through electrostatic interaction between peptidyl guanadinium groups and phosphate in the NA backbone. Stereospecific hydrogen bonding holds free arginine to NA codonal nucleobases. Several plausible configurations could exist for the NA/peptide/amino acid complex: the amino acid and peptide could be bound to the same strand, they could be bound to separate stands or one or both could be bound to both strands. Formation of Watson-Crick bonds between the two NA strands moves arginine to a position that permits formation of a peptide bond between it and the extant peptide. These NA/peptide and NA/amino acid stereospecific interactions, combined with template-based NA replication, provide the basis for the homochirality of sugars and amino acids in biological molecules.
Energy for peptide bond formation in our model’s translation is provided by autocatalytic energy transfer from phosphorylated peptidyl guanidinium groups. Thus, peptide elongation does not involve aminoacylation as in modern translation but rather uses phosphate bond energy directly as in the ATP-driven formation of peptide bonds in glutathione (Janowiak and Griffith 2005). When arginine-enriched peptides are not already present, translation can be initiated by formation of a bond between arginine’s amine group and the activated carboxyl group of an amphiphilic organic acid (aromatic amino acid, neutral peptide, amphiphilic multimer or fatty acid). The bond energy for this initial ‘peptide’ bond derives from the reactive amphoteric acid (acyl phosphate or thioester) formed through environmental photochemical or volcanic processes. This aspect of the model ensures that base-enriched peptides are situated at the oil/water emulsified interface and can keep NAs there.
Our model protobiont has no ribosomes to serve as the site for translation. However, arginine-enriched peptides assume some ribosomal structural and functional roles—binding to the phosphate backbones of NAs, holding them together and catalyzing bond energy transfers that allow peptide bond formation. NAs not directly involved in translation are also bound to the base-enriched peptides, providing a stable structural platform for translation.
A Specific Testable Candidate for the Protobiont
Here we present a specific oligonucleotide dimer for the core arginine translator of our model protobiont. Clearly this NA can be readily synthesized and its functional interactions with arginine and arginine-enriched peptides (i.e. the collective ability to replicate, bind arginine to NA, incorporate arginine into peptides and promote phosphate transfer) can be tested through controlled experiments. Although we believe this specific candidate NA is a reasonable first step in developing an experimental protocol to test the hypothesis of NA/peptide interdependence, we recognize it may prove to be partially or even wholly incorrect. Certainly other related or independent models could be developed and tested.
Based on the following arguments, our model protobiont’s core NA is the palindromic DNA hexamer 5′AGCGCT. In our scenario biological function began under hyperthermal conditions so that DNA rather than RNA is favored. A palindrome is suggested because of simplicity and efficiency in replication and translation. I chose a six base strand length as a compromise between speed and fidelity of replication (favoring short NAs) and binding affinity for arginine and peptides (favoring greater length). Furthermore, internal bonds will cause looping with the ends stuck together in longer palindromes. The middle four bases are guanine and cytosine to provide tighter binding during translation and copying under hot conditions. In order to incorporate all currently used nucleotide bases, the terminal bases are adenine and its complement, thymine. The sequence 5′AGCGCT has the arginine codon CGC as well as its anticodon. It also bears the first two bases (AG) of the second motif for arginine’s six codons (plausibly serine could have co-opted the AGC codon from arginine during the evolution of the universal genetic code as it seems to have later co-opted AGG and AGA in mitochondria [Osawa et al. 1989]). Based on experimental evidence (Knight and Landweber 1998; Yarus et al. 2005), the codonal sequences could provide specificity for arginine. Recognition and orientation of arginine by NA strands bearing its codon and anticodon provide an evocative model for how the genetic code started.
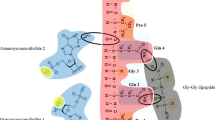
Figure 1 depicts how two strands of the palindromic hexamer 5′AGCGCT might bind arginine and facilitate its incorporation into a peptide during translation. It should be noted that the long hydrocarbon skeleton of arginine provides enough flexibility for the phosphorylated guanadinium group of a terminal arginine residue to interact with its own free carboxyl group, permitting peptide bond formation. Basic amino acids with shorter hydrocarbon chains (in keeping with the length of all other amino acids except lysine) will not work in our model. Of course, the long bodies of arginine and lysine also facilitate versatile interactions with NA.
Translation of an arginine-enriched peptide by the nucleic acid 5′AGCGCT. A strand of nucleic acid (1) is bound to an arginine-enriched “peptide” (4–7) through phosphate-guanadinium electrostatic bonds. Hydrophobic residues on this “peptide” (7) hold the complex at the aqueous-lipid interface (not shown). A second strand (2) holds free arginine (3) via stereospecific hydrogen bonding. Formation of Watson-Crick bonds between complementary strands places the free arginine in a position where a peptide bond can form with the activated C-terminal arginine residue (4). Energy for peptide bond formation is provided by the phosphorylated guanidium group of this C-terminal arginine residue
Direction of Subsequent Evolution of Living Systems
A detailed account of the subsequent evolution of life—from the simple protobiont we have modeled to extant organisms—is beyond the scope of this paper. However, a brief overview of some key aspects of early molecular evolution is appropriate. The first significant development allowing the evolution of complex extant living systems was an increase in translation capability through expansion of the genetic code. This, in turn, allowed the evolution of metabolic pathways able to provide energy, synthesize building blocks and support cellular organization. As living systems proliferated and evolved, the oil slick and environmental supplies of building blocks and energy-rich compounds would become exhausted. The substrate depletion model for the retrograde development of metabolic pathways (Horowitz 1945) is a plausible mechanism during the early stages of biochemical evolution. Increased biological sophistication through cellular organization would allow membrane-based phosphorylation using proton gradients and development of photosynthetic pathways, originally based upon pigments within the oil slick rather than within cells.
The expansion of the genetic code requires a source of NA variability and selective pressures favoring certain NA sequences over others. Woese (1998) has proposed a compelling scenario in which the early development of the genetic code entailed massive pre-organismal lateral gene exchange. In our model the protobiont includes not only the core arginine translator that provides a local milieu favoring NA aggregation, replication and modification, but a global pool of other NA strands and peptides. Some of the NAs will have properties that are of value to a developing living system like binding useful amino acids and/or incorporating them into peptides; some preformed peptides will have useful properties as well. It is not unreasonable that reverse translation (as well as reverse transcription) could occur under these circumstances, dramatically accelerating molecular evolution.
Under our molecular interdependence scenario, natural selection favors aggregates of mutually beneficial NAs that collectively use environmental resources most effectively for growth and reproduction. Thus, the original arginine-translating palindrome becomes supplemented and then replaced with linear mRNAs coding for diverse complex and efficient peptides and by a growing battery of “tRNAs” specific for different amino acids. In the earliest stages these “tRNAs” would recognize amino acids through stereospecific binding (albeit likely not of high affinity) accounting for the observed binding affinity of many amino acids to RNA aptamers bearing their anticodons (Yarus et al. 2005). Our scenario provides an explanation for the seemingly paradoxical correlation of RNA binding sites with codons for arginine but anticodons for other amino acids. As the translation repertoire grew, tRNAs and adaptor enzymes (homologous with extant aminoacyl tRNA synthetases, able to recognize amino acid and tRNA and catalyze aminoacylation at the 3′ adenosine of tRNA) would replace direct stereospecific recognition of amino acids at the codon or anticodon.
Some evidence supports the so-called “co-evolution theory for the genetic code” which proposes codonal assignments were determined through evolution of amino acid biosynthetic pathways (Wong 1975; DiGiulio 2008). It seems reasonable that adaptor enzymes could function catalytically in their cognate amino acid’s synthesis as well as serving to recognize tRNA and catalyze aminoacylation. Consequently, the expansion of the genetic code could indeed be related to biosynthetic pathway evolution, but it may have been through retrograde development, terminal steps first. In light of our premise of molecular interdependence, the earliest amino acids incorporated into the genetic code would be of compelling utility to the evolving living system and may therefore have been complex, not simple ones.
References
Arnez JG, Augustine JG, Moras D, Francklyn CS (1997) The first step of aminoacylation at the atomic level in histidyl-tRNA synthetase. Proc Natl Acad Sci USA 94:7144–7149
Bada JL (2004) How life began on Earth: a status report. Earth Planet Sci Lett 226:1–15
Barbier B, Visscher J, Schwartz AN (1993) Polypeptide-assisted oligomerization of analogs in dilute solution. J Mol Evol 37:554–558
Benner SA, Cohen MA, Gonnet GH (1993) Reading the palimpest: contemporary biochemical data and the RNA world. In: Gesteland RF, Atkins JF (eds) the RNA world. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Bernstein M (2006) Prebiotic materials from on and off the early Earth. Phil Trans R Soc B 361:1689–1702
Cairns-Smith G (1982) Genetic takeover and the mineral origins of life. Cambridge University Press, Cambridge
Cavalier-Smith T (1987) The origin of cells: a symbiosis between genes, catalysts and membranes. Cold Spr Hbr Symp Exp Biol 52:805–824
Cech TR, Zaug AJ, Grabowski PJ (1981) In vitro splicing of the ribosomal RNA precursor of Tetrahymena: Involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell 27:487–496
Chyba C, Sagan C (1992) Endogenous production, exogenous delivery and impact-shock synthesis: an inventory for the origins of life. Nature 355:125–132
Copley SD, Smith E, Morowitz HJ (2005) A mechanism of association of amino acids with their codons and the origin of the genetic code. Proc Natl Acad Sci USA 102:4442–4447
Crampton DJ, Guo S, Johnson DE, Richardson CC (2004) The arginine finger of bacteriophage T7 gene 4 helicase: role in energy coupling. Proc Natl Acad Sci USA 101:4373–4378
Crick FHC (1968) The origin of the genetic code. J Mol Biol 38:367–379
Day W (1984) Genesis on planet Earth. Yale Univ Press, New Haven CT
Deamer DW (1997) The first living systems: a bioenergetic perspective. Microbiol Mol Biol Rev 61:239–261
Deamer D, Singaram S, Rajamani S, Kompanichenko V, Guggenheim S (2006) Self assembly processes in the prebiotic environment. Phil Trans R Soc B 361:1809–1818
DeDuve C (1991) Blueprint for a cell: the nature and origin of life. Neil Patterson, Burlington, NC
DiGiulio M (2008) An extension of the coevolution theory of the evolution of the genetic code. Biol Direct 3:37
Doudna JA, Cech TR (2002) The chemical repertoire of natural ribozymes. Nature 418:222–228
Eschenmoser A (2004) The TNA-family of nucleic acid systems: properties and prospects. Orig Life Evol Biosp 34:277–306
Ferris JR (2006) Montmorillonite-catalysed formation of RNA oligomers: the possible role of catalysts in the origin of life. Phil Trans R Soc B 361:1777–1786
Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M (1996) RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enitioselectivity. Nucleic Acids Res 24:1029–1036
Gilbert W (1986) The RNA world. Nature 319:618
Griffith RW (1994) The life of the first vertebrates. Bioscience 44:408–417
Hagen WJ Jr, Parker A, Steuerwald A, Hathaway M (2007) Phosphate solubility and the cyanate-mediated synthesis of pyrophosphate. Orig Life Evol Biosp 37:113–122
Hochachka PW, Somero GN (1984) Biochemical adaptation. Princeton University Press, Princeton, NJ
Horowitz NH (1945) On the evolution of biochemical synthesis. Proc Natl Acad Sci USA 31:153–157
James KD, Ellington AD (1997) Surprising fidelity of template-directed chemical ligation of oligonucleotides. Chem Biol 4:595–605
Janowiak BE, Griffith OW (2005) Glutathione synthesis in Streptococcus agalachae: one protein accounts for gamma-glutamylcysteine synthetase and glutathione synthetase activities. J Biol Chem 280:11829–11839
Jay DG, Gilbert W (1987) Basic protein enhances the incorporation of DNA into lipid vesicles: model for the formation of primordial cells. Proc Natl Acad Sci USA 84:1978–1980
Jeffares DC, Poole AM, Penny D (1998) Relics from the RNA world. J Mol Evol 46:18–36
Joyce GF (2002) The antiquity of RNA-based evolution. Nature 418:214–221
Joyce GF, Orgel L (1999) Prospects for understanding the origin of the RNA world. In: Gesteland RF, Cech TR, Atkins JF (eds) The RNA world, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Knauth AP (1998) Salinity history of the Earth’s early ocean. Nature 395:554–555
Knight RD, Landweber LF (1998) Rhyme or reason: RNA-arginine interactions and the genetic code. Chem Biol 5:R215–R220
Knight R, Landweber L, Yarus M (2003) Tests of a stereochemical genetic code. In Lapointe J, Brakier-Gingras L (eds) Translation mechanisms. Springer
Koonin EV (2007) The cosmological model of eternal inflation and the transition from chance to biological evolution in the history of life. Biol Direct 2:15
Kunin V (2000) A system of two polymerases—a model for the origin of life. Orig Life Evol Biosp 30:459–466
Lahav N (1991) Prebiotic coevolution of self-replication and translation or RNA world? J Theor Biol 151:531–539
Lahav N (1993) The RNA-world and coevolutionary hypotheses and the origin of life: implications, research strategies and perspectives. Orig Life 23:329–344
Lasaga AC, Holland HD, Dwyer MJ (1971) Primordial oil slick. Science 174:53–55
Lazcano A, Fox GE, Oro J (1992) The origin and evolution of early Archean cells. In: Mortlock RP (ed) The evolution of metabolic function. CRC, Boca Raton, FL
Line MA (2002) The enigma of life and its timing. Microbiology 148:21–27
Lunine JI (2006) Physical conditions on the early Earth. Phil Trans R Soc B 361:1721–1731
Maizels N, Weiner AM (1987) Peptide-specific ribosomes, genomic tags and the origin of the genetic code. Cold Spring Hbr Symp Quant Biol 52:743–749
Mason SF (1991) Chemical evolution: origin of the elements, molecules and living systems. Clarendon, Oxford
Miller SL (1987) Which organic compounds could have occurred on the early Earth? Cold Spring Hbr Symp Quant Biol 52:17–27
Morowitz HJ, Kostelnik JD, Yang J, Cody GD (2000) The origin of intermediary metabolism. Proc Natl Acad Sci USA 97:7704–7708
Muirhead H (1987) Pyruvate kinase. In: Jurnak FA, McPherson A (eds) Biological macromolecules and assemblies: Active sites of enzymes. vol 3. Wiley, New York
Nancollas GH, Tomazik B (1974) Growth of calcium phosphate on hydroxyapatite crystals: effect of supersaturation and ionic medium. J Phys Chem 78:2218–2225
Nashimoto M (2002) The RNA/protein symmetry hypothesis: experimental support for reverse translation of primitive proteins. J Theor Biol 209:181–187
National Research Council (2007) The limits of organic life in planetary systems. National Academies, Washington DC
Nelson KE, Levy M, Miller SL (2000) Peptide nucleic acids rather than RNA may have been the first genetic material. Proc Natl Acad Sci USA 97:3868–3871
Nilson FPR (2002) Possible impact of a primordial oil slick on the atmospheric and chemical evolution. Orig Life Evol Biosp 32:247–253
Nisbet EG, Sleep NH (2001) The habitat and nature of early life. Nature 409:1083–1091
Orgel LE (1968) Origin of the genetic apparatus. J Mol Biol 38:381–393
Orgel LE, Lohrman R (1974) Prebiotic chemistry and nucleic acid replication. Acc Chem Res 7:368–377
Osawa S, Ohama T, Jukes TH, Watanabe K (1989) Evolution of the mitochondrial genetic code. I. Origin of AGR serine and stop codons in metazoan mitochondria. Mol Evol 29:202–207
Prinn RG, Fegley B Jr (1989) Solar nebula chemistry: origin of planetary, satellite and cometary volatiles. In: Atreya SK, Pollack JB, Mathews MS (eds) Origin and evolution of planetary and satellite atmospheres. University of Arizona Press, Tucson
Reeve JN, Bailey KA, Li W-T, Marc F, Sandman K, Soares DJ (2004) Archaeal histones: structures, stability and DNA binding. Biochem Soc Trans 32:227–230
Rittenger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ (1997) Structure at 165 Å of RhOA and its GTP-binding protein in complex with a transition state analog. Nature 389:758–762
Rosen R (1984) Relational biology and the origin of life. In: Matsuno K, Dose K, Harada K, Rohlfing DL (eds) Molecular evolution and protobiology. Plenum, New York
Segré D, Ben-Eli D, Deamer DW, Lancet D (2001) The lipid world. Orig Life Evol Biosp 31:119–145
Shanks AL (1983) Surface slicks associated with tidally-forced internal waves may transport pelagic larvae of benthic invertebrates and fishes shoreward. Mar Ecol Prog Ser 13:311–315
Shapiro R (1999) Prebiotic cytosine synthesis: a critical analysis and implications for the origin of life. Proc Natl Acad Sci USA 96:4396–4401
Shapiro R (2006) Small molecule interactions were central to the origin of life. Quart Rev Biol 81:105–125
Shock E (1992) Chemical environments of submarine hydrothermal systems. Orig Life Evol Biosp 22:67–107
Sleep NH, Zahnle KJ, Kasting JF, Morowitz HJ (1989) Annihilation of ecosystems by large asteroid impacts on the early Earth. Nature 342:139–142
Stribling R, Miller SL (1987) Energy yields for hydrogen cyanide and formaldehyde syntheses: the HCN and amino acid concentrations in the primitive ocean. Orig Life Evol Biosp 17:261–273
Tervahattu H, Tuck A, and Vaida V (2004) Chemistry in prebiotic aerosols: a mechanism for the origin of life. In Seckbach J (ed) Cellular origin and life in extreme habitats and biology. Springer Netherlands
Tian F, Toon OB, Pavlov AA, DeSterck HA (2005) Hydrogen-rich early Earth atmosphere. Science 308:1014–1017
Trifonov EN (2004) The triplet code from first principles. J Biomol Struct Dyn 22:1–11
Urey HC (1952) On the early chemical history of the Earth and the history of life. Proc Natl Acad Sci USA 38:351–363
Usher DA (1977) Early chemical evolution of nucleic acids: a theoretical model. Science 196:311–313
Wächtershauser G (1988) Before enzymes and templates: theory of surface metabolism. Microbiol Rev 52:452–484
Wächtershauser G (1992) Groundworks for an evolutionary biochemistry: the iron-sulfur world. Prog Biophys Mol Biol 58:85–201
Wächtershauser G (2000) Life as we don’t know it. Science 289:1307–1308
Wächtershauser G (2006) From volcanic origins of chemoautotrophic life to Bacteria, Archaea and Eukarya. Phil Trans R Soc B 361:1787–1808
Weiner AM, Maizels N (1987) Transfer RNA—like structures tag the 3’ ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci USA 84:7383–7387
Woese CR (1967) The genetic code: the molecular basis for gene expression. Harper and Row, NY
Woese CR (1968) The fundamental nature of the genetic code: prebiotic interactions between polynucleotides and polyamino acids or their derivatives. Proc Natl Acad Sci USA 59:110–117
Woese CR (1998) The universal ancestor. Proc Natl Acad Sci USA 95:6854–6859
Wolf YI, Koonin EV (2007) On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation and subfunctionalization. Biol Direct 2:14
Wong JT (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Wood TD, Guan Z, Borders CL Jr, Chen LH, Kenyon GL, McLafferty FW (1998) Creatine kinase: essential arginine residues at the nucleotides binding site identified by chemical modification and high-resolution tandem mass spectrometry. Proc Natl Acad Sci USA 95:3362–3365
Yakhnin AV (2007) A model for the origin of protein synthesis as coreplicational scanning of nascent RNA. Orig Life Evol Biosp 37:523–536
Yarus M, Caporaso JG, Knight R (2005) Origins of the genetic code: the escaped triplet hypothesis. Annu Rev Biochem 74:179–198
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Griffith, R.W. A Specific Scenario for the Origin of Life and the Genetic Code Based on Peptide/Oligonucleotide Interdependence. Orig Life Evol Biosph 39, 517–531 (2009). https://doi.org/10.1007/s11084-009-9169-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-009-9169-2