Abstract
During the RNA World, organisms experienced high rates of genetic errors, which implies that there was strong evolutionary pressure to reduce the errors’ phenotypical impact by suitably structuring the still-evolving genetic code. Therefore, the relative rates of the various types of genetic errors should have left characteristic imprints in the structure of the genetic code. Here, we show that, therefore, it is possible to some extent to reconstruct those error rates, as well as the nucleotide frequencies, for the time when the code was fixed. We find evidence indicating that the frequencies of G and C in the genome were not elevated. Since, for thermodynamic reasons, RNA in thermophiles tends to possess elevated G+C content, this result indicates that the fixation of the genetic code occurred in organisms which were either not thermophiles or that the code’s fixation occurred after the rise of DNA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to the RNA World hypothesis, the genetic code evolved in primordial organisms with RNA-based genomes (Szathmary 1999; Freeland et al. 1999). The genetic machinery in those organisms can be assumed to have been highly prone to errors. Therefore, these organisms experienced selective pressures to reduce the phenotypical impact of errors. There is strong evidence that as a result of those pressures, the genetic code evolved a structure that increased the probability that errors lead to synonymous or near-synonymous codons (Sonneborn 1965; Woese 1965; Haig and Hurst 1991; Freeland and Hurst 1998; Freeland et al. 2000; Gilis et al. 2001; Freeland et al. 2003).
It is clear that the more frequent a type of error was, the greater should have been the selective evolutionary pressure it exerted on the still evolving code. Concretely, we here hypothesize that the amount of adaptation to any particular type of error was in proportion to its primordial rate of occurrence, in line with prior heuristic arguments (Ardell 1998). Once the genetic code was fixed it stayed virtually unchanged until the present. This means that the structure of the standard genetic code today should still reflect the relative prevalence of the different kinds of genetic errors of the time when the code was fixed. Indeed, for example G→A errors should have been more frequent than G→C errors (just as they are today), according to a simple model of polymerase errors which should apply to primordial polymerases (Topal and Fresco 1976). Because the former errors exerted more evolutionary pressure on the code, the code should have become better adapted to minimize their impact. This is in fact the case, as is straightforward to read off from the structure of the code.
Our aim is to systematically exploit the proportional correlation between the structure of the standard code and primordial evolutionary pressures in order to draw quantitative conclusions about the primordial organisms in which the genetic code was shaped. To this end, we will statistically examine the structure of the known standard genetic code in order to reconstruct to some extent the rates of the genetic errors that shaped the code. This will also provide some information about the nucleotide frequencies of the genomes of the primordial organisms of the time when the genetic code was fixed.
Formalizing the Error-reduction Hypothesis
We begin by formulating our error-reduction hypothesis mathematically. First, let π X denote the frequency of each nucleotide in the genome, where X can take the four values U,C,A,G (in RNA). These were in part determined by the environment. For instance, it is known that π G + π C tends to be elevated in RNA of thermophiles because G and C share a triple bond that stabilizes RNA at high temperatures (Galtier and Lobry 1997). Such a temperature dependence of π G + π C is to be expected in a primordial RNA world. Next, we denote the twelve (conditional) probabilities of “point” substitution errors by M XY , where X is the original nucleotide and Y is the new nucleotide. These probabilities represent the primordial error rates of either the replication or the translation machinery, whichever error rates were exerting more evolutionary pressure on the genetic code. Our analysis applies in either case.
We also introduce for every substitution X→Y a variable T XY which quantifies the average “tolerance” of the genetic code to the substitution X→Y. Concretely, we let T XY be the average value of an experimentally-obtained measure of chemical proximity of pairs of amino acids. To calculate T XY we average over all pairs that can be obtained by a substitution X→Y. T XY is large when the amino acid after the substitution tends to be identical or chemically similar to the amino acid before the substitution. Notice that T XY depends not only on the chemical relatedness of amino acids but also on the detailed structure of the genetic code. We also introduce for each nucleotide X a variable σ X which quantifies its “safety,” defined as the average value of amino acid chemical proximity whenever an X occurs and is substituted by any other nucleotide. σ X is obtained from the T XY values by averaging over the three possible substitutions of X. For the precise formula of T XY and σ X we have used, see the “Materials and Methods” section below.
According to the above error-reduction hypothesis the amount of tolerance that the evolving assignment of codons to amino acids developed for a particular type of genetic error was in proportion to the prevalence of this type of error (for possible evolutionary mechanisms, see (Knight et al. 2001a,b)). Let us, therefore, consider any two substitutions X →Y and I →J, with uneven absolute probabilities π X M XY and π I M IJ , respectively. For instance, if X →Y is the more frequent substitution, i.e., if π X M XY > π I M IJ , then the hypothesis requires that the genetic code should have preferentially adapted to the substitutions X →Y. Therefore, the tolerance value T XY should be greater than the tolerance value T IJ .
In addition, also the varying nucleotide frequencies should have exerted correspondingly varying evolutionary pressures. To see this, assume, for instance, that a nucleotide X was more abundant in the genome than a nucleotide Y. In this case, even if X and Y were equally error-prone, there was more evolutionary pressure for the genetic code to evolve so as to tolerate errors in X than there was pressure for the code to evolve to tolerate errors occurring in Y, because in this case the absolute probability of error is higher for X. The outcome of the evolution of the genetic code should therefore have been such that when a nucleotide X is substituted by some other nucleotide then the resulting amino acid is on average more similar to the original amino acid than when a nucleotide Y is substituted. What we called the safety value, σ X , measures this tolerance: it is the average chemical proximity of the pair of amino acids resulting from any substitution of X. Therefore, according to our hypothesis, π X > π Y led to σ X > σ Y . (This argument does of course not assume that all nucleotides were equally error prone. σ X is the average chemical proximity conditional on X getting substituted, which means that σ X is largely insensitive to the nucleotides being unequally error prone.)
Materials and Methods
In order to describe the relations between the σ X values and the π X values, as well as in between T XY and π X M XY , we use the following equations. For the error rates, we postulate that differences in the error rates led to the evolution of proportional differences in the tolerances:
This is a linear approximation and as such will be valid as long as the differences among tolerance values are small. In Eq. 1, the right hand side quantifies the evolutionary pressure due to errors, while the left hand side quantifies the error-reducing adaptation of the genetic code. The proportionality parameter, F m , expresses the strength of the coupling between the evolutionary pressure and the adaptive response of the genetic code. For example, a large F m would mean that the coupling is strong, namely that the code’s adaptation to the various types of errors depends very sensitively on the relative prevalence of these errors.
Similarly to the error rates, we postulate the following proportionality law relating differences in nucleotide frequencies to differences in safety values
where F π is a constant. The precise relationship may be nonlinear but this linear approximation should be very good since the values of π X − π Y are likely to have been small. Finally, we note that π X and M XY satisfy two further equations, namely the probabilistic consistency condition ∑ π X = 1 and the definition of an overall error rate ε = ∑ Y ≠ X π X M XY . We set ε = 1% but the precise value of ε is immaterial because multiplying it by a constant c causes no change except multiplying all the M XY values by c. The values of F m and F π are estimated below. As is shown in the Appendix, the values of F π and F m can be chosen in a wide range around the best estimate without significantly affecting the conclusions.
In regards to the tolerance and safety values, there is considerable literature on measuring the overall fitness of a genetic code, see e.g. Gilis et al. (2001). Here, we need to quantify in more detail the fitness of the code with respect to individual types of error. To this end, we first note that the number of occurrences of each nucleotide X in the genetic code is 64 ·3/4 = 48. We label these occurrences by k = 1,..,48 and the corresponding codons by C k (a codon that contains multiple X’s is labeled multiple times). We can then define the tolerance value, T XY , through:
Eq. 3 contains a sum over all k in order to take into account that the X which is being substituted can be in any one of the 48 codons C k . Further, \( EX\left[A(C_k),A(\widehat{C_k})\right] \) is the so-called “Experimental Exchangeability” (EX) value (Yampolsky and Stoltzfus 2005). It is an experimentally obtained quantitative measure of how efficient a resulting protein is, on average, if a substitution error replaced a codon, C k , by another codon, \( \widehat{C_k} \), i.e., it measures the chemical proximity of the amino acids A(C k ) and \( A(\widehat{C_k}) \). We chose to use EX because it was obtained from enzyme efficiency data, rather than from data on non-synonymous substitutions, which means that EX is not contaminated with the structure of the standard genetic code. For problems in this respect with the PAM matrices, see Di Giulio (2001). By definition, \( EX\left[A(C_k), A(\widehat{C_k})\right] = 0 \) when the amino acids are most dissimilar and the value is 1 if the amino acids are identical, i.e., if the codons C k and \( \widehat{C_{k}} \) are synonymous. Note that due to the averaging in Eq. 3, our results will be only very weakly dependent on the details of the amino acid proximity measure. We extended EX to include stop codons, and set EX = 1 for all errors between two stop codons and EX = 0 whenever an amino acid is replaced by a stop codon or vice versa. This extension may underestimate the effect of those errors on proteins but the resulting inaccuracy in T XY is small because, as before, each T XY consists of a large number of terms. Indeed, work by Jestin and Kempf (1997) indicates that the assignment of stop codons is more closely related to deletion errors than to point substitutions.
Equation 3 also weighs each term by the probability, r k , for codon C k . Namely, let us denote in codon C k the remaining two nucleotides a(k) and b(k). Assuming that on average the probability of a codon is the product of the probabilities of its nucleotides, then the probability, r(k), that when X occurs it does so in codon C k , is given by r(k) = π a(k) π b(k). Finally, Eq. 3 contains weight factors, p t(k), which take into account the fact that the probability of a substitution may also depend on the position of X in the codon. Namely, let t(k) = 1,2,3 denote the position of X in the codon C k . If a substitution possesses unequal probabilities, p t , of occurring at codon positions t = 1,2,3, then the contribution of a substitution X→Y to T XY needs to be weighted by the probability, p t(k), for its position, t, in the codon. If the genetic code is adapted mainly to reduce the effect of mutations then there cannot be any position bias, i.e., we have \( (p_1, p_2, p_3) = (\frac{1}{3}, \frac{1}{3}, \frac{1}{3}) \). If, however, the genetic code is adapted mainly to reduce the effect of mistranslations then some bias towards errors in the first and third positions is likely (Parker 1989), such as \( (p_1, p_2, p_3) = (\frac{5}{16}, \frac{1}{16}, \frac{10}{16}) \). The latter triplet p k was used in calculating Fig. 1. Our reconstructed nucleotide frequencies vary only very little, about 1% , between this bias triplet and the no-bias triplet, as is explained in the robustness analysis section below.
The safety value σ X of any nucleotide X is found by taking the weighted sum of the tolerance values for the three possible substitutions of X. The weights are given by the probabilities of each substitution, i.e.,
where Y 1, Y 2, Y 3 are the other three nucleotides and \( N_X = M_{XY_1} + M_{XY_2} + M_{XY_3} \) is the normalization constant. Note that due to this normalization the varying error-proneness of nucleotides has little effect on the safety values σ X . Moreover, of the four nucleotides, Cytosine is likely to be the most error-prone (due to spontaneous deamination), and therefore our method would tend to slightly overestimate the value of π C in the primordial genome. Because we find that the G + C content is not elevated, this slight bias only re-enforces the conclusion.
Finally, we note that among the 12·11 equations of the form Eq. 1 generically 11 are linearly independent. Similarly, of the 4 ·3 equations of the form Eq. 2, generically 3 are independent. Therefore, Eqs. 1 and 2 together with ∑ π X = 1 and ε = ∑ Y ≠ X π X M XY are solvable for the 16 variables π X and M XY after we estimate the two values of F m and F π . To this end, we note that for typical values of π X and M XY we obtain values of T XY − T IJ ≤ 250 and σ X − σ Y ≤ 200. On average, π X M XY = ε/12 and π X = 25% , which indicates that corresponding to the two ranges above are π X M XY − π I M IJ ≈ ε/24 and π X − π Y ≈ 12.5% . Therefore, for the computation we set F m = 250/(ε/24) and F π = 200/0.125. As will be shown in Appendix, our results are robust against variations in the estimated values of F m and F π .
Results and Discussion
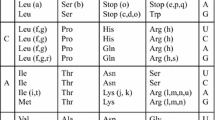
The results of solving Eqs. 1 and 2 for the unknowns are displayed in Fig. 1. In particular, Fig. 1a shows the reconstructed values of the primordial nucleotide frequencies. We find that π G + π C = 50.1% , i.e., the G + C content is not elevated. This result supports, therefore, the hypothesis that the genetic code evolved in non-thermophile organisms. This finding fits a scenario proposed by Poole et al. (1999), Forterre and Philippe (1999) and others in which adaptations for high temperatures seen in some modern taxa are comparatively late evolutionary developments, rather than primordial features. Given that the Last Universal Common Ancestor (LUCA) was likely a thermophile (Schwartzman and Lineweaver 2004), we propose that there was a change in the biosphere after the fixation of the standard code: While the primordial organisms were non-thermophile, later organisms leading towards LUCA evolved adaptations to high temperatures.
Reconstructed nucleotide frequencies, a, and reconstructed absolute probabilities of errors, b, at the time the genetic code stopped evolving. The rates of forward and backward substitutions were found to be similar and each column in b displays the larger of the two rates, because its value is known with more certainty as discussed in the text. The scale of the bars in b linearly scales with ε – an assumed overall error rate (here ε = 1% ), but ε affects neither the relative rates of errors nor the nucleotide frequencies
Also, notice that Fig. 1a reports only the content of the coding parts of the genome, because errors in non-coding regions exert no selective pressures on the genetic code. Independently of whether or not such non-coding regions existed in the early genomes, if the primordial organisms had been thermophiles, we should still have found elevated G + C in the coding regions because elevated G + C content is necessary for the heat stability of these regions of the genome. Also, interestingly, the G + C levels in modern organisms are dependent on the codon position (Muto and Osawa 1987). We are here not pursuing this further but we note that most of the position dependence of the base frequencies is unrelated to environmental influences, such as the temperature, and that it is instead mostly due to the position-dependent evolutionary pressure imposed by the genetic code (Freeland et al. 2001). Indeed, the G+C frequencies at the three positions are highly correlated, i.e., they tend to be elevated jointly (Muto and Osawa 1987).
An alternative but less probable scenario is that the code underwent extensive error-reducing evolution after the adoption of DNA-based genomes. Because there is no correlation between temperature and G + C content for DNA, our results would then be consistent with the analysis of the genetic code by Di Giulio (2003) who argues for a thermophile or hyperthermophile origin of the code. However, it is likely that the evolution of the genetic code took place in RNA-based organisms and was mostly complete by the time DNA-based organisms evolved. This is because DNA-based organisms had much longer genomes (Penny et al. 2003) and their length precludes significant changes in the structure of the genetic code (see also Gutfraind 2006)
Aside from the nucleotide frequencies, we reconstructed the (absolute) error rates, π X M XY (Fig. 1b). Interestingly, we find that the rates of transition errors were consistently high, in line with nucleotide substitution rates of modern organisms (cf. Li 1996). We note also that the reconstructed error rates are consistent with rates of replication errors that are to be expected in a double-stranded genome, where the rates of errors should approximately equal the rates of their complements. This is indeed approximately the case (see Fig. 1b).
Finally, we note that, because of the way the structure of the genetic code reduces the impact of errors, it is not possible with our method to fully reconstruct the differences between the rates of forward and backward substitutions. This is because when an error replaces an amino acid A 1 with an amino acid A 2 that is similar, then the error in the opposite direction also tends to lead to a chemically similar amino acid. Thus, if for a genetic code a tolerance value T XY is elevated then for that genetic code the tolerance value T YX for the reverse substitution also tends to be elevated. As a result, the reconstructed values π X M XY and π Y M YX tend to be close even if these rates were not as close when the genetic code was fixed. The more frequent of the two errors likely had the most influence in shaping the genetic code, thus “shadowing” the lesser error. A detailed analysis of this “shadowing” and of other sources of uncertainty is described in the Appendix. There, it is shown that even under extreme assumptions our value for π G + π C increases to merely about 56%. Thus, our main conclusion that the genetic code was fixed in organisms that were not thermophilic or not in an RNA world is robust. More generally, further analysis along the lines of this paper may yield interesting additional insights about the primordial environment or properties of the early genetic machinery.
References
Ardell D (1998) On error minimization in a sequential origin of the standard genetic code. J Mol Evol 47(1):1–13
Di Giulio M (2001) The origin of the genetic code cannot be studied using measurements based on the PAM matrix because this matrix reflects the code itself. J Theor Biol 208:141–144
Di Giulio M (2003) The universal ancestor was a thermophile or a hyperthermophile: tests and further evidence. J Theor Biol 221:425–436
Forterre P, Philippe H (1999) Where is the root of the universal tree of life? BioEssays 21:871–879
Freeland S, Hurst L (1998) The genetic code is one in a million. J Mol Evol 47:238–248
Freeland S, Knight R, Landweber L (1999) Do proteins predate DNA? Science 286(5440):690–692
Freeland S, Knight R, Landweber L, Hurst L (2000) Early fixation of an optimal genetic code. Mol Biol Evol 17(4):511–518
Freeland, S, Knight, R, Landweber, L (2001) A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol 2(4), research0010.1–0010.13
Freeland S, Wu T, Keulmann N (2003) The case for an error minimizing standard genetic code. Orig Life Evol Biosph 33:457–477
Galtier N, Lobry J (1997) Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J Mol Evol 44:632–636
Gilis D, Massar S, Rooman M (2001) Optimality of the genetic code with respect to protein stability and amino-acid frequencies. Genome Biol 2(11), research0049.1–0049.12
Gutfraind A (2006) Error-tolerant coding and the genetic code. Master’s thesis, University of Waterloo
Haig D, Hurst L (1991) A quantitative measure of error minimization in the genetic code. J Mol Evol 33:412–417
Jestin J-L, Kempf A (1997) Chain termination codons and polymerase-induced frameshift mutations. FEBS Lett 419:153–156
Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP (2001) RNA-catalyzed RNA-polymerization: accurate and general RNA-templated primer extension. Science 292:1319–1325
Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD (2005) Exploration of factors driving incorporation of unnatural dNTPS into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase A. Nucleic Acids Res 33(8):2620–2628
Knight R, Freeland S, Landweber L (2001a) Rewiring the keyboard: evolvability of the genetic code. Nat Rev Genet 2:49–58
Knight R, Landweber L, Yarus M (2001b) How mitochondria redefine the code. J Mol Evol 53:299–313
Li W-H (1996) Molecular evolution. Sinauer, Massachusetts
Muto A, Osawa S (1987) The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A 85, 166–169
Parker J (1989) Errors and alternatives in reading the universal genetic code. Microbiol Rev 53(3):273–298
Penny D, Hendy MD, Poole AM (2003) Testing fundamental evolutionary hypotheses. J Theor Biol 223(3):377–385
Poole A, Jeffares D, Penny D (1999) Early evolution: prokaryotes, the new kids on the block. BioEssays 21:880–889
Schwartzman D, Lineweaver C (2004) The hyperthermophilic origin of life revisited. Biochem Soc Trans 32(2):168–71
Sonneborn TM (1965) Degeneracy of the genetic code: extent, nature, and genetic implications. In: Bryson V, Vogel HK (eds) Evolving genes and proteins. Academic, New York
Szathmary E (1999) The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet 15(6):223–229
Topal M, Fresco J (1976) Complementary base pairing and the origin of substitution mutations. Nature 263:285–289
Woese CR (1965) On the evolution of the genetic code. Proc Natl Acad Sci U S A 54:1546–1552
Yampolsky L, Stoltzfus A (2005) The exchangeability of amino acids in proteins. Genetics 170(4):1459–1472
Acknowledgements
We are indebted to Jean-Luc Jestin and Rob Knight for valuable comments and Henry Wolkowicz for advise on computational methods. This work has been supported by the Natural Sciences & Engineering Research Council of Canada, CFI, OIT, PREA and the Canada Research Chairs Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Materials Original data and programs are available at the author’s web site: http://www.math.uwaterloo.ca/~akempf/gc/.
Appendix
Appendix
Robustness Analysis
We verified numerically that for codes significantly different from the actual genetic code, our method would predict lower or higher G+C contents, as the case may be. In the following, we show that our result, namely that the G+C content is average, is nevertheless robust. We divide our analysis into four points. In points 1–3 of this robustness analysis, F m and F π are set to the estimated values described in the “Materials and Methods” section. In point 4 we address the robustness of the results with respect to changes in F m and F π .
-
(1)
Regarding the “shadowing” effect, we expect that strong shadowing of the rates of transitions would have the largest impact on our data because transitions make the largest contributions to the calculation of the safety values, σ X . In particular, if the rates of the transitions U→C and A→G are shadowed, then the value of π U + π A would be systematically elevated, and the value π G + π C would be systematically suppressed. This is unlikely to have been the case because σ G is found to be quite low (ultimately because amino acids where G is in the second position have relatively low degeneracy) while σ U is quite high (because of relatively high degeneracy). We can put an upper bound on the impact of shadowing on the nucleotide frequencies by considering the case of a larger than possible shadowing. To this end, we set the values of T UC and T AG at 50% of normal. We found that even in this extreme case π G + π C increases to merely 56.7%. Such a small elevation of π G + π C is still far below what is expected in a thermophile (Galtier and Lobry 1997).
-
(2)
Equations 1 and 2 are linear approximations to what are likely non-linear equations of the form π X M XY − π I M IJ = f(T XY − T IJ ) and π X − π Y = g(σ X − σ Y ). Fortunately, the linear approximation can be assumed to be very good because the set of non-linear f and g that are consistent with the error-reduction hypothesis is very restricted. Recall that π X M XY > π I M IJ must lead to T XY > T IJ , and likewise, π X > π Y must imply σ X > σ Y . Therefore,f and g must be monotonically increasing functions. Thus, the ranking of the error rates and nucleotide frequencies are determined by the values of tolerance and safety alone, which are themselves largely fixed once the structure of the genetic code is known. Moreover, the differences in the tolerance and safety values are small, making the linear approximations more accurate. Note that Eqs. 1 and 2 do in fact already incorporate some non-linearity in the sense that r(k) in T XY depends non-linearly on the nucleotide frequencies, while σ X also depends non-linearly on M XY .
One test for the impact of non-linearity is to replace Eq. 2 with \( (\sigma_C - \sigma_Y)^2 = \widehat{F_\pi} (\pi_C - \pi_Y) \) where C is Cytosine and Y stands for any of the three other nucleotides. This choice satisfies the considerations above because after calculating we find that indeed σ C > σ Y and π C > π Y for all Y. Here, \( \widehat{F_\pi} \) is the value of the proportionality constant adjusted for the non-linearity (\( \widehat{F_\pi} = 200^2/0.125 \)). Solving this modified equation (simultaneously with the unchanged Eq. 1) leads to less than 1% change in nucleotide frequencies and less than 0.2% change in π G + π C .
-
(3)
The effect of the position bias of errors on our results is very small. In order to see this, we model this bias by the triplet p t as described in the “Materials and Methods” section. Choosing no position bias instead of the non-uniform bias also described in the “Materials and Methods” section and used in Fig. 1 leads to a change of at most 1% in the nucleotide frequencies. This insensitivity is plausible: a frequently-occurring nucleotide would have caused the genetic code to structure in such a way that errors in this nucleotide are less harmful no matter in which position in a codon the nucleotide occurs. There is a stronger dependence on the position bias in the reconstructed rates of error. In particular, one finds a greater gap between the rates of transitions and the rates of transversions in the biased case. This difference is especially noticeable when F m is small. The effect is a natural consequence of the structure of the genetic code: the code is particularly adapted to transition errors in the third position, as can be seen directly by examining how the degeneracies are placed in the code.
-
(4)
The dependence of our results on the values of F π and F m is plotted in Fig. 2. Notice that over the entire range of possible values of the parameters F π and F m the sum π G + π C ranges merely between 50 and 52% . Thus, our conclusion that the G + C content is not significantly elevated is robust with respect to varying the values of F π and F m . Regarding the determination of the values of F π and F m , we notice on the one hand that, for large values of F π and F m , the curves in Fig. 2 converge. This is because if in Eq. 2, for instance, σ X − σ Y is known and F π is doubled, then the difference π X − π Y is halved. The value of the parameter F m affects the solution in a similar way. Regardless of the value of F m , transitions are always more frequent than transversions. This is nicely consistent with a simple model that may apply to primordial polymerases (Topal and Fresco 1976), according to which higher rates of transitions than transversions are to be expected due to the chemical differences between pyrimidines and purines. We note that while the sources of polymerase errors in modern organisms are rather more complex, see Kincaid et al. (2005), transitions are more frequent than transversions even in modern organisms (Li 1996). Also, it will be very interesting to compare our reconstructed error rates to the fidelity of RNA-based polymerases once their general statistics has been established. For first experiments in this direction, see (Johnston et al. 2001). In our calculation, exactly how different the two types of rates are depends of course on the value of F m . For instance, when F m = 250/(ε/10) the ratio between the rates of transitions and transversions is between 3 and 4 (cf. Li 1996).
Reconstructed nucleotide frequencies, a, and absolute probabilities of errors, b, when the genetic code stopped evolving, as a function of the parameters F π and F m , respectively, down to the minimum possible values of F π and F m . a shows that the reconstructed value of π G + π C in the primordial genome is very close to 50% regardless of the value of F π . Thus, our finding that π G + π C was not elevated is robust. b shows how the rates of transition errors (top four curves) and the rates of transversion errors (bottom eight curves) vary with F m .
On the other hand, the parameters F π and F m also cannot be arbitrarily small since else the equations would not possess solutions. For instance, a very small value of F π for a fixed σ X − σ Y would require the difference π X − π Y to be so large that the constraint that the sum of probabilities is one (∑ π X = 1) no longer holds. F m cannot be very small for similar reasons, as this would be inconsistent with the overall error rate being equal to ε. An estimate of the lower bound on F m follows from the size of the gap between the rates of transitions and the rates of transversions in modern organisms. With the exception of mitochondria, the average rate of transitions is rarely more than four times the average rate of transversions (Li 1996; Parker 1989). This corresponds to F m ≥ 250/(ε/10), which is indeed well below the best estimate value that we used above, namely 250/(ε/24).
Rights and permissions
About this article
Cite this article
Gutfraind, A., Kempf, A. Error-reducing Structure of the Genetic Code Indicates Code Origin in Non-thermophile Organisms. Orig Life Evol Biosph 38, 75–85 (2008). https://doi.org/10.1007/s11084-007-9071-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-007-9071-8