Abstract
Following the near-obliteration of American chestnut (Castanea dentata [Marsh.] Borkh.) by the chestnut blight early in the last century, interest in its restoration has been revived by efforts to develop a blight-resistant form of the species. We summarize progress and outline future steps in two approaches: (1) a system of hybridizing with a blight-resistant chestnut species and then backcrossing repeatedly to recover the American type and (2) transformation of American chestnut with a resistance-conferring transgene followed by propagation and conventional breeding. Several decades of effort have been invested in each approach. More work remains, but results indicate that success is within practical reach. The restoration of C. dentata to its native habitat now appears to be less a matter of time and conjecture than ever before in 90 years of work by public and private entities. The difficult and protracted task of incorporating extraspecific genes for resistance into a tree species with lethal susceptibility to a naturalized pathogen represents perhaps the most extreme of restoration challenges. Its pursuit by a small non-governmental organization supported primarily by philanthropy and volunteers may serve as a model for other species threatened by exotic pathogens or insects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the most serious contemporary ecological issues are those arising from the introduction of pathogens and other pests to new plant hosts. Such introductions, usually inadvertent consequences of international trade and travel, can be devastating when the new host has few natural defenses and the new pest few natural controls. A well-known example is that of the chestnut blight pathogen (Cryphonectria parasitica) and American chestnut (Castanea dentata [Marsh.] Borkh.). Following the introduction of the blight to North America from Asia sometime prior to 1904, virtually every one of billions of mature American chestnut trees was killed during the first half of the twentieth century (Gravatt 1949; Hepting 1974; Newhouse 1990). At present, the species persists in the wild state almost exclusively as shade-suppressed seedlings or small basal sprouts from blight-killed trees (Griffin et al. 1983; Stephenson et al. 1991; Paillet 2002; Dalgleish et al. 2016). Extremely few American chestnuts have survived infection for extended periods as large trees.
Following the invasion of a new forest pest, some fraction of the tree population typically escapes or is genetically resistant, and the species persists in a reduced state, perhaps to recover at some future time. American chestnut represents the rare case in which the pathogen is pervasive (it survives on other hosts) and lethal susceptibility is so nearly universal that the species may be rescuable only by incorporating extraspecific alleles or genes through interspecific hybridization or genetic transformation. Early attempts to recover American chestnut focused on hybridizing C. dentata with Asian species of the genus, which carry genes for blight resistance. Two breeding programs were initiated in the 1920s: one by the United States Department of Agriculture (USDA) and one by the Brooklyn Botanic Garden, later transferred to the Connecticut Agricultural Experiment Station (CAES) (Burnham et al. 1986; Anagnostakis 2012). Breeding work in both programs stopped in the early 1960s, and neither succeeded in reaching its objective of combining the blight resistance of Asian chestnut with the growth and form of American chestnut (Jaynes 1978; Burnham et al. 1986).
A new strategy for breeding blight-resistance into C. dentata was proposed by Charles Burnham, a maize geneticist, beginning in the early 1980s (Burnham 1981, 1988; Burnham et al. 1986). Burnham and others created The American Chestnut Foundation (TACF) for the purpose of raising funds for the project, and breeding activities were consolidated on land in Meadowview, Virginia, in 1989. In a separate line of work, the New York Chapter of TACF began collaborating in 1990 with the SUNY College of Environmental Science and Forestry (SUNY-ESF) to enhance blight resistance in American chestnut by transgenesis.
The breeding work by TACF and its collaborators is still underway. This program, and a smaller one described by Griffin (2006), have been the only sustained programs of breeding a blight-resistant American chestnut since the 1960s. The long-anticipated completion of the breeding plan elaborated by Burnham et al. (1986) is very close in two (of many) breeding populations. Despite the fact that few people alive today actually have a memory of American chestnut as a forest tree, reports of progress by TACF and its collaborators have renewed widespread interest in restoring the species, a task that could become the largest reintroduction effort ever undertaken for a plant species (Jacobs et al. 2013). Our purpose here is twofold: (1) to evaluate actual progress in the breeding program and outline future steps, including integration of the breeding and transgenic work, and (2) to summarize the work of TACF as perhaps a paradigmatic example of in extremis species restoration by a not-for-profit private organization.
Backcross breeding to incorporate blight resistance
The Burnham plan
Based upon an analysis of published and unpublished data from the USDA and CAES programs, Burnham et al. (1986) reasoned that alleles for blight resistance could be transferred to the C. dentata genome by creating a hybrid with Chinese chestnut (C. mollissima Bl.) or other blight-resistant Castanea species and then repeatedly backcrossing to C. dentata while selecting for resistance alleles. Selection for resistance alleles would be possible, even when they are necessarily heterozygous after each backcross to C. dentata, because alleles for blight resistance were found to be partially dominant in the earlier programs, and there appeared to be two genes involved. Each generation of backcrossing to C. dentata would halve the C. mollissima fraction of the genome, on average, thus facilitating recovery of the C. dentata type in all respects except susceptibility to chestnut blight. The full breeding plan called for the production of a third backcross (B 3) that would be intercrossed to create a segregating B 3 F 2 population in which a fraction of the trees would be homozygous for all resistance alleles from the original Asian parent. Finally, following testing and selection for blight-resistance, the B 3 F 2 plantation would become a seed production orchard for a B 3 F 3 generation that would be essentially C. dentata in character but sufficiently blight-resistant to begin restoration of the species. Other details of the breeding plan are provided by Hebard (2006) and Jacobs et al. (2013).
As implemented at Meadowview beginning in 1989, the Burnham plan was expedited by beginning with two first-backcross (B 1) hybrid trees from the USDA and CAES breeding programs, called the ‘Clapper’ and ‘Graves’ trees, respectively. Each was derived from an F 1 hybrid between C. dentata and C. mollissima that was subsequently backcrossed to C. dentata (Hebard 2006). It was assumed that the Clapper and Graves trees might not contain identical alleles for blight resistance, and accordingly they have been bred in different breeding populations as distinct sources of resistance.
For the Clapper and Graves sources of blight resistance, a minimum of 25 sets of C. dentata trees have been used as parents in each backcross generation to enhance the capture of American allelic diversity and minimize inbreeding in the final, intercrossing generation (Namkoong 1991; Hebard 2002). An operational assumption was that as many as three unlinked loci carry resistance alleles, which meant that as few as one in 64 trees in the B 3 F 2 generation were expected to be homozygous for resistance at all loci. To ensure the highest, practical probability of recovering at least nine homozygous individuals per American line, the Clapper and Graves B 3 F 2 test plantations (before conversion to seed production orchards through selection) were designed to contain 1350 offspring from each open-pollinated B 3 line, or a total of 33,750 trees for a resistance source with 25 American lines (Hebard 2002). Because of the size of the task, these plantations have taken many years to build.
Progress in breeding the Clapper and Graves sources in the Meadowview breeding program
Breeding began by crossing the Clapper and Graves B 1 trees with 66 and 76 C. dentata individuals, respectively, with an average yield of 12 B 2 seedlings per cross. Selected Clapper and Graves B 2s were crossed with 52 and 43 additional C. dentata individuals, with an average yield of 37 seedlings per cross. To evaluate resistance, B 2 and B 3 trees were inoculated in the spring of their fifth growing season with strains of C. parasitica with low (SG2-3) and high (Ep155) pathogenicity but both capable of killing C. dentata sprouts. We have found subsequently that resistance to the two strains is strongly correlated genetically (r g = 0.95 ± 0.07), suggesting that common loci confer resistance to both. However, using both strains extends the range of response levels detectable in a test. All individuals were culled except those with the smallest cankers for each strain. Additional selection was performed for C. dentata morphological traits among individuals with the smallest cankers (Hebard 1994), leaving approximately 4% of B 2 and B 3 trees to be used in further breeding. After selection in the B 3, grand-progeny remained from 29 of the 66 C. dentata trees crossed with Clapper and 25 of the 76 trees crossed with Graves, exceeding the target of 20 American lines for each resistance source.
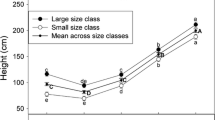
Assuming blight resistance is controlled by three incompletely dominant genes at segregating loci, and assuming successful selection for heterozygosity at all three loci, the B 2 and B 3 trees used for further breeding should have resembled F 1 hybrids in having intermediate levels of resistance. The average canker sizes or ratings of B 2 and B 3 selections met these expectations (Fig. 1). Furthermore, Diskin et al. (2006) found that the American phenotype was recovered in 96% of B 3 trees as judged by 24 scored or measured traits typically used to distinguish C. dentata from C. mollissima. These findings provided evidence that the breeding program was working as planned.
Average blight canker areas (A) or average visual canker ratings (B) of selected Clapper and Graves B 2s (A) and B 3s (B) relative to C. mollissima and C. dentata controls and the F 1 hybrid between the two species. Each tree was inoculated in separate locations on the stem with both a weakly (SG2-3) and highly pathogenic (Ep155) strain of C. parasitica. Letters above bars within a panel indicate significant differences (P < 0.05, Tukey HSD tests) after accounting for inoculation year as a covariate
Intercrosses to generate B 3 F 2s were carried out with open pollination among the B 3 selections. Creation of the Clapper and Graves B 3 F 2 test plantations at Meadowview began in 2002 and continues. As of 2015, 20 of 29 Clapper American lines and 13 of 25 Graves lines had been planted in all 9 blocks of their respective plantations (Table 1). Preliminary selection for blight resistance in the plantations is carried out at 2 years of age by artificially inoculating stems with the SG2-3 strain of C. parasitica. Typically, 80–85% of the trees are culled within a year after artificial inoculation because of significant canker expansion. At present, further selections are made after longer-term response to the initial inoculation, response to additional inoculations or naturally occurring infections, and progeny-testing.
Approximately 10,000 seedlings remained in the Clapper and Graves B 3 F 2 test plantations at Meadowview in 2015, and 11,250 had not yet been planted (Table 1). The plantations will not become completed seed orchards until the Clapper plantation is reduced to approximately 260 trees with the highest resistance, and the Graves plantation similarly reduced to approximately 225 trees. Although B 3 F 3 seeds have been distributed from these plantations for testing purposes since 2008, their blight resistance has necessarily been limited by progress in selection (Table 1). In field trials in forest settings, progeny from B 3 F 2 trees have grown significantly faster than C. mollissima seedlings and not significantly slower than C. dentata (Clark et al. 2016). As of 2016, the trees in these trials are not yet large enough to support epidemics of blight, so their “field resistance” remains unknown.
Genetic variation in blight resistance after preliminary selection in the B 3 F 2
Experimental field tests of half- or full-sib progeny sets are the proof of parental genetic quality in a breeding program. Beginning in 2009, TACF has been progeny-testing B 3 F 2 trees that have passed the initial phenotypic selection for blight resistance, and to date we have completed evaluations of B 3 F 3 progenies from 293 Clapper and 131 Graves B 3 F 2 seed parents in experimental plantations at Meadowview. The seed parents were open-pollinated with a random and presumably average sample of other B 3 F 2 trees shedding pollen. Castanea dentata and C. mollissima, the progeny of intraspecific crosses, were also included in progeny tests as blight-susceptible and resistant controls. The progenies are challenged in their third growing season with weakly (SG2-3) and strongly (Ep155) pathogenic strains of the C. parasitica.
These progeny tests have provided the first estimates of heritability and the first experimental proof of progress toward the projected culmination of the plan presented by Burnham et al. (1986). Figure 2 shows the composite results of these tests using best linear unbiased predictions (BLUP) of mean responses to both strains of the pathogen.
Severity of cankering in open-pollinated progeny sets from 293 Clapper and 131 Graves B 3 F 2 mother trees and C. mollissima and C. dentata controls following inoculation by weakly (SG2-3) and highly pathogenic (Ep155) strains of C. parasitica. Raw data for the Tukey box plots are best linear unbiased predicted mean scores for approximately 12 B 3 F 3 offspring per seed parent. Scores were scaled to a mean of 0 for C. mollissima and a mean of 100 for C. dentata. The dashed line shows the expected mean response to inoculation (49.8) for the offspring of a hypothetical seed orchard thinned to the most resistant of the 424 seed parents. Estimation of actual genetic gain following parental selection is based on the formula Gain = 2h 2 S, where h 2 is the heritability (narrow sense) for family means estimated from the progeny tests (0.45), and S is the selection differential. In this example, we assumed that the selected parents would be those whose progeny means were at least 1.6 standard deviations better than the mean for all B 3 F 3 trees
Based on the assumptions of the breeding program, the expected average resistance level of all B 3 F 3 offspring from unselected B 3 F 2 parents is that of an F 1 population, i.e., intermediate between C. dentata and C. mollissima as in Fig. 1. However, even after initial phenotypic selection for resistance among B 3 F 2 trees, the average level of cankering severity in B 3 F 3 offspring is more similar to C. dentata than to C. mollissima (Fig. 2). This result suggests that some selections in the B 2 or B 3 generations did not inherit the full set of blight resistance alleles from the source C. mollissima. Every offspring from backcrosses to C. dentata had at least a 50% complement of susceptibility alleles from the American parent, and most had more. The object, apparently not always achieved, was to identify those trees that had exactly a 50% complement of resistance alleles from C. mollissima. Additional evidence of imperfect selection is the fact that we found significant (P < 0.001) heritable variation in canker size among Clapper and Graves B 3 F 2 half-sib families. Assuming random pollination of the seed parents, there should have been no genetic variation in blight resistance among B 3 F 2 families if selected B 3 trees were heterozygous at all resistance loci.
The most likely explanation for these results is that some resistance alleles were lost through backcrossing to C. dentata. This would certainly have happened if resistance is controlled by more than three loci, in which case it would have been statistically improbable to recover all resistance alleles in every American line given the size of populations that could feasibly be generated and evaluated in field trials. Nonetheless, with the number of American lines across which the breeding program is replicated (25 and 29 for the two sources of resistance), we believe it is unlikely that the breeding program has completely lost any resistance alleles from the Clapper and Graves ancestors. If true, this means that resistance can be improved through further breeding.
Variation among B 3 F 3 progeny sets (Fig. 2) is the result of genetic differences among individual B 3 F 2 seed parents, some inevitably more blight-resistant than others, that were open-pollinated by a random host whose genetic quality was average. In other words, even the most resistant B 3 F 3 families were limited by the mediocrity (on average) of their many male parents. This limitation may have been further constrained by the tendency of heavily blighted trees to produce excessive numbers of male catkins. The average genetic resistance of B 3 F 3s will increase until all but the most blight-resistant B 3 F 2s are culled in the process of converting the test plantations to seed orchards, at which time both seed and pollen parents will be members of a selected population. Currently, the mean B 3 F 2 breeding values for canker severity are closer to C. dentata than to C. mollissima. Selection of the 5% most resistant B 3 F 2 from the 10,000 remaining trees is predicted to yield B 3 F 3 progeny with average canker severity that is almost exactly (by coincidence) intermediate in resistance between the average C. mollissima and the average C. dentata (Fig. 2). Actual gains may be less depending upon the level of genotype by environment interaction. Considering that the most resistant B 3 F 2s have breeding values closer to C. mollissima than to C. dentata within pollen clouds from partially selected plantations, the maximum resistance of the B 3 F 3 progeny is expected to approach the mean resistance of C. mollissima upon completion of selection at B 3 F 2. Restoration trials composed of B 3 F 3 progeny from selected B 3 F 2 parents will determine whether resistance after selection at B 3 F 2 is sufficient to generate reproductive, self-sustaining populations in the forest. To accelerate crown closure and help ensure success over competing vegetation, restoration plantings will typically have densities far greater than can survive to maturity (Nyland 1996), so it will not be necessary that all trees have high resistance.
Progress in breeding additional sources of resistance
In addition to the Clapper and Graves sources of resistance, TACF’s Meadowview program has advanced 22 other sources of Asian chestnut resistance variously from the F 1 through B 4 (Table 2). The purpose of this work is to increase the diversity of resistance alleles in the breeding program and to minimize vulnerability to evolution of virulence in the fungus (Hebard 2004). Castanea mollissima has been the preferred source of blight resistance because canker expansion after inoculation with Cryphonectria parasitica was slower on C. mollissima than on other Castanea species (Graves 1950; Clapper 1952). Two other Asian species of chestnut (C. crenata and C. seguinii Dode.) have also been utilized. Some of the C. mollissima sources of blight resistance (‘Nanking’, ‘Meiling’, and ‘Kuling’) were highly blight-resistant selections from the USDA’s breeding program for nut production (McKay and Jaynes 1969).
It is still not known whether different C. mollissima trees carry different alleles for resistance. This was investigated at Meadowview by test-crossing each of 16 C. mollissima trees with three others and inoculating the progeny at age 3 with the highly pathogenic Ep155 strain of C. parasitica. Additive genetic variance in the offspring—which would arise if individual parents differed in their average contribution to resistance—was not significantly greater than zero (likelihood ratio test P = 0.17). However, non-additive genetic variance associated with specific crosses was significant (P = 0.002). Non-additive genetic variance in blight resistance (e.g., variance resulting from dominance or epistasis) cannot be “fixed” to incrementally enhance resistance across generations, but it can contribute to the population mean level of resistance after selection in a given generation.
State chapter breeding programs
As a complement to the breeding program at Meadowview, the Burnham plan has been replicated by volunteers in most of TACF’s 16 state and multi-state chapters beginning with the Pennsylvania Chapter in 1994. In addition, members of the New York Chapter of TACF have been the principal sponsors of the SUNY-ESF work to enhance blight resistance through genetic transformation of C. dentata. The transformation project will be discussed in a subsequent section.
Most of the chapter activities are based on the Meadowview model and were started with pollen from selected B 2s and B 3s created at Meadowview using Graves or Clapper sources of resistance. There are 14 of these programs (Table 3). Chapter breeding efforts serve the important purpose of ensuring regional climatic adaptability in the future restoration program by using locally autochthonous C. dentata trees in backcross generations after the B 2.
A second, related purpose of the chapter-directed programs is to increase the diversity of the C. dentata genome captured by the breeding program. As shown by Hebard (2012), the contributions of the chapter breeding programs to the inbreeding effective population size of the anticipated founding population are expected to be substantial (>200 in aggregate). For these reasons, the chapter programs are regarded as critical to the task of restoring the species. These programs are assisted by four regional coordinators who are full-time employees of TACF, but the actual breeding work depends almost entirely on volunteer leadership and labor. Almost half of the state chapters have started creating B 3 F 2 or B 4 F 2 test plantations, and the earliest may be completely planted (though not culled to the final selections) as early as 2020. Chapter breeding programs manage 586 plantations of breeding material on approximately 335 ha of public and private land not owned by TACF, and volunteers log over 50,000 h annually in this effort (Table 4, also see Fig. 5 in Jacobs et al. 2013). Considering the scope, complexity, and duration (20+ years) of the chapter breeding projects, this application of citizen science and volunteer labor to the restoration of a species is exceptional and perhaps unique among conservation programs.
Genetic mapping and genomic selection for blight resistance
All published genetic maps of blight-resistance loci in chestnut are based on a population of F 2 progeny from a cross between two F 1 hybrids involving C. dentata and the ‘Mahogany’ cultivar of C. mollissima (Table 2). The ‘Mahogany’ cultivar is the Chinese ancestor for the Graves source of resistance. Kubisiak et al. (1997) constructed a genetic map of Castanea comprising 184 molecular genetic markers in 12 linkage groups corresponding to the haploid chromosome number for chestnut. Three major quantitative trait loci (QTLs) for resistance were positioned on three different linkage groups and accounted for about 40% of the phenotypic variation in canker size. This finding bolstered an earlier decision to structure the breeding program to accommodate three genes for blight resistance (Hebard 1994). The initial map by Kubisiak et al. was later expanded twice, first with 189 markers (Sisco et al. 2005) and lastly with 447 markers (Kubisiak et al. 2013) along with an integrated physical map (Fang et al. 2012). In addition, the three major blight resistance QTLs have recently been sequenced with candidate genes further resolved (Staton et al. 2015).
However, we are not confident that we know the number of loci conferring resistance to the blight in C. mollissima. The population in which blight resistance QTLs were mapped was small (n = 85). Loci with minor effects may have been undetected, and effect sizes of significant loci may have been inflated (Beavis 1994). Furthermore, published maps are based on just one source of resistance, the cultivar ‘Mahogany’, out of over 20 sources of resistance that are currently being advanced (Table 2) and that could differ in their resistance-conferring loci.
It would be desirable to progeny-test all ca. 7000 candidates (Table 1) for final retention in the B 3 F 2 orchards, but this would be an enormous and expensive task. Short of this, we believe that genomic selection (Meuwissen et al. 2001) may be the most effective alternative to selection based strictly on the phenotypes of inoculated B 3 F 2 trees. Genomic selection consists of genotyping a training population of progeny-tested trees with a genome-wide panel of genetic markers and then estimating marker effects on the breeding values of trees as measured by the performance of their progeny. These effects are then used to estimate the breeding values of the complement population of genotyped but not-progeny-tested trees. Modeled simulations theoretically indicate that the efficiency of tree breeding can be radically improved by genomic selection (Grattapaglia and Resende 2011). Our own simulations using parameters specific to the TACF breeding program indicate that genomic selection can markedly speed completion of the B 3 F 2 seed orchards and do so at a substantially reduced cost compared with progeny testing. Simulations assumed a training population of 500 trees per source of resistance, each genotyped at 20,000 single nucleotide polymorphisms. Under this approach, 90% of all culling decisions in the B 3 F 2 would be based on phenotype, 2% on progeny testing, and 8% on genomic selection. However, we shall remain cautious about this approach until its operational efficacy and practicality are demonstrated (Grattapaglia and Resende 2011).
Enhancing blight resistance through genetic transformation
Over nearly the same timespan as the backcross breeding program has developed, the SUNY-ESF group has independently pursued a transgenic approach to enhancing blight resistance in C. dentata, supported by philanthropy and volunteer assistance channeled through TACF. One of the authors (Powell) is a lead scientist within the SUNY-ESF group. The first advance in this line of research was the successful establishment of C. dentata somatic embryo cultures at the University of Georgia (Merkle et al. 1991). A completed transformation system was fully developed 16 years into the program (Polin et al. 2006; Maynard et al. 2006), and the first transgenic C. dentata trees were planted in field trials in 2006 under a permit from the US Department of Agriculture, Animal Plant Health Inspection Service (USDA-APHIS).
The gene for oxalate oxidase (OxO) has become the lead gene for this transgenic research. The OxO gene is nearly ubiquitous in cereals and is present in some other monocots and many dicots including strawberry, beet, peanut, and apricot, but not Castanea. Oxalate production by some species of plant pathogenic fungi, including C. parasitica, facilitates their pathogenicity by killing plant cells in advance of invading fungal hyphae (Hebard et al. 1984; Hebard and Shain 1988; Kim et al. 2008). The OxO gene detoxifies oxalate. Because decreased oxalate production in C. parasitica is associated with reduced virulence (Havir and Anagnostakis 1983, 1985; Chen et al. 2010), it was hypothesized that detoxifying oxalate at the canker margins would protect plant cells and enhance resistance to the blight. A natural effect of OxO is to augment host resistance, and numerous studies have shown that overexpressing OxO (i.e., producing more of the enzyme or producing it in additional tissues) in transgenic plants can enhance resistance to a variety of pathogens (Donaldson et al. 2001; Liang et al. 2001; Schneider et al. 2002; Hu et al. 2003; Livingstone et al. 2005; Dong et al. 2008; Walz et al. 2008; Partridge-Telenko et al. 2011; He et al. 2013; Zhang et al. 2013; Newhouse et al. 2014).
The SUNY-ESF group has created a number of transgenic C. dentata events that express OxO, where “event” refers to a single transformed cell that is then clonally multiplied in cultures and regenerated into whole plants. These plants exhibit enhanced resistance to C. parasitica (Zhang et al. 2013; Newhouse et al. 2014) (Fig. 3). Several of these events are being tested for non-target effects (e.g., D’Amico et al. 2014), and no differences have been found to date between transgenic and non-transgenic plants.
Results of small-stem assay 30 days after inoculation with C. parasitica strain EP155. A Wild-type C. dentata cell line ‘Ellis 1’. All wilted between 19 and 30 days. B Transgenic event ‘Darling 54’ produced from ‘Ellis 1’ and expressing the oxalate oxidase transgene. None wilted within 30 days and all survived with small healing cankers at the inoculation sites, from which C. parasitica could be isolated. The plants were still surviving after 10 months. C Tissue cultured clone from the ‘Qing’ variety of C. mollissima. Half wilted between 24 and 30 days and only one eventually survived
More recently, the SUNY-ESF group and others have begun studying the possible use of transgenes from Asian chestnut species. Transgenes from related species are sometimes referred to as cisgenes. Twenty-seven candidate cisgenes have been identified based upon differential gene expression at canker margins between C. mollissima and C. dentata, proximity to QTLs for blight resistance, or presumed functions based on similar genes in other plants (Baier 2010; Barakat et al. 2012; Nelson et al. 2014). These candidate genes have been used to create genetically transformed C. dentata somatic embryos. Preliminary results indicate enhanced pathogen resistance in events overexpressing several known genes. None has demonstrated resistance as high as that conferred by the OxO gene, nor is it expected that any single gene from C. mollissima will confer full resistance to C. parasitica.
Next steps
Breeding
With accelerated progress using genomic selection for blight resistance, we expect that final culling in the Clapper and Graves test plantations at Meadowview will be finished by 2022, thus completing the first full implementation of the Burnham plan. Controlled crosses are currently being made among the most blight-resistant of the progeny-tested B 3 F 2s, and their progeny will provide us with an early indication of the resistance levels that will be realized from the completed seed orchards. The crosses may yield individuals with excellent resistance, in which case elite families with reduced genetic diversity could be developed for plantings where high, fairly uniform resistance is desired.
Current evidence (Fig. 2) suggests that the completed Clapper and Graves B 3 F 2 seed orchards will yield progeny with an average resistance that is less than that of C. mollissima. Genetic gains that are greater than projected in Fig. 2 might be achieved by eliminating entire American lines that appear to be significantly inferior because they have lost one or more resistance alleles. Although a useful fraction of progeny from the completed orchards might be sufficiently blight-resistant for restoration, the Meadowview breeding plan has been conservatively revised to incorporate recurrent selection for blight resistance in additional generations (B 3 F 3, B 3 F 4, etc.) beginning with random mating within the Clapper and Graves seed orchards. Simulations indicate that genetic gains in a plan like this could be further increased through a form of full-sib family selection based on the use of markers to reconstruct pedigrees in open-pollinated populations (El-Kassaby and Lstiburek 2009; Lstiburek et al. 2015). Recurrent selection will produce populations with an increasingly larger fraction of highly resistant individuals in each subsequent generation, and an average level of resistance within the range of C. mollissima may be reached by the B 3 F 4 or B 3 F 5.
The Clapper and Graves resistance lines will be combined beginning with the B 3 F 3 generation, and the breeding population will be further augmented with additional resistance sources (Table 2). Aside from presumably enhancing the genetic basis of blight resistance, these additions to the recurrent breeding line will also augment C. dentata genetic diversity within the breeding population and increase the inbreeding effective population size. The generation time for recurrent selection will probably be at least 10 years.
When fully developed, genomic selection is expected to expedite the resource-intensive process of evaluating blight resistance. For new sources of resistance, selection for C. dentata portions of the genome not involved in resistance may enable the program to bypass the third backcross generation without sacrificing progress toward the C. dentata type.
With baseline resistance established by the breeding program, it is hoped that self-sustaining populations of C. dentata could become established with natural selection. Because of the thinning that inevitably occurs within a naturally regenerated or planted stand of trees, it will not be necessary for all or even a majority of seedlings in a restoration population to have high blight-resistance. Stocking models for upland hardwoods indicate that no more than about 25% of planted trees will survive natural thinning to become a mature stand of pure chestnut, even at relatively light initial densities (Gingrich 1967). In practice, even fewer chestnut trees would survive thinning because other, volunteer species typically become part of the final stand in upland hardwood reforestation projects. Mixed stands of upland hardwoods, including C. dentata, are the natural and desirable state. The ultimate goal of the breeding program is a set of chestnut populations that are regionally adapted, have sufficient blight resistance to survive, and have enough genetic variability to evolve further on their own.
Transformation
Two OxO transformation events of C. dentata, ‘Darling 54’ and ‘Darling 58’, will be submitted in 2017 for regulatory review by USDA-APHIS, the US Environmental Protection Agency, and the US Food and Drug Administration. The Darling 54 and Darling 58 events occurred in ramets of the same clone of C. dentata (‘Ellis 1’), so they should be identical in all respects other than the transformation event itself. During the review process, the transgenes from these two events will be bred into one or more seedling populations of C. dentata within the restrictions imposed by the review process. The reason for this is that chestnuts typically do not grow well when propagated from tissue culture or other clonal means, but seedling populations carrying the transgenes can be created in a single generation by selecting for the 50% of the offspring that are transgenic. Trees carrying the OxO transgene can be rapidly identified using an enzyme assay. Additional breeding could be pursued without further review if non-regulated status is approved by USDA-APHIS and a registration granted by the EPA. A second generation of breeding would fix the transgene in the homozygous state in about one-fourth of the intercrossed progeny.
Assuming regulatory approval, it is expected that seed from these trees will be released to the public for private use, and the work of breeding the transgene into seedling populations of C. dentata will be continued by TACF. Private use would not necessarily include large-scale restoration of C. dentata to natural forest lands. The use of transgenic C. dentata to restore the species would require breeding the transgene into large, regionally adapted populations such as those of the state chapter backcross breeding programs. Resistant transgenic trees could also be used effectively to rescue allelic diversity among surviving C. dentata by controlled or open pollination with flowering trees that occasionally develop from long-suppressed seedlings and sprouts after the canopy is disturbed. Whether TACF will pursue these strategies is uncertain at this time, and in part this depends upon whether tests show that C. dentata carrying the OxO transgene is competitive under wild conditions.
Discussion
Resistance to C. parasitica even in Asiatic species is not absolute. Four of the authors (Hebard, Fitzsimmons, Powell, Steiner) observed the regular occurrence of C. parasitica cankers on C. mollissima in apparently wild stands in China. Identification was confirmed by isolation and culture of the fungus. In wild populations we found that the cambium was generally uninfected, as indicated by the absence of exposed xylem. We also observed blight cankers causing deformation of trees cultivated in nut orchards, with symptoms similar to those observed on C. mollissima in the U.S. (Jones et al. 1980). Chestnut blight is regarded in China as an important disease in orchards (Tarcali and Radocz 2009; Yan et al. 2007).
These facts suggest that even if all the alleles for blight resistance in C. mollissima were completely fixed within C. dentata, blight likely would continue to be a problem on trees that are severely weakened by competition or adverse environments. Circumstances that favor the vigorous growth of C. dentata may be critical to successful restoration in forest stands. Hypoviruses and other parasites of C. parasitica (Milgroom and Cortesi 2004) could act synergistically with genetic blight resistance to render more complete disease control than can be achieved by either factor acting alone.
We have not addressed a second problematic pathogen of American chestnut, Phytophthora cinnamomi. This non-native oomycete causes a root disease that is frequently fatal to chestnut and many other tree species in the warmer portions of the C. dentata distribution, especially on heavy clay soils in the Piedmont region of the southeastern United States (Crandall et al. 1945). Research by TACF and partnering institutions has shown that some C. mollissima trees are genetically resistant to P. cinnamomi, including the ‘Mahogany’ ancestor of the Graves resistance source and many of its descendants in the TACF breeding program (Jeffers et al. 2012; Zhebentyayeva et al. 2013). Resistance to P. cinnamomi within Graves appears to be controlled by alleles with major effects. In the absence of previous selection for resistance to P. cinnamomi, a subset of progeny-tested Graves B 3 F 2s has resistance that approaches that of C. mollissima controls (Joseph B. James and others, unpublished). We believe that resistance to P. cinnamomi and C. parasitica could be combined though controlled crosses between B 3 F 2 trees carrying the greatest resistance to either pathogen, and then performing recurrent selection at B 3 F 3 and B 3 F 4 for resistance to both pathogens. Resistance to both pathogens could also be achieved by breeding Phytophthora-resistant B 3 F x trees with blight-resistant transgenic trees.
The prolonged but eventually abortive breeding projects by USDA and CAES seem to have put a damper on American chestnut breeding for the next couple of decades. By the 1970s, when forest tree breeding was at its apogee in the U.S. (Wheeler et al. 2015), there was virtually no active breeding for blight resistance in American chestnut, and informed opinion was pessimistic about the future of the species (Hepting 1974). In the judgment of one prominent forest geneticist of the time, neither the federal government nor private industry would undertake the difficult process of breeding a blight-resistant American chestnut after the failures of previous decades (J. W. Wright, personal communication to Steiner, 1970). That statement has proven to be remarkably prescient during the nearly 50 years since, but it did not anticipate the potential role of a non-profit corporation and enthusiastic scientists and laypersons.
Beginning in the early 1990s, TACF’s breeding program has revived optimism that American chestnut can be restored to its former habitats, and this growing optimism appears to have catalyzed a great deal of scientific interest in the species. This is shown in part by the fact that research activity on this species, which has been ecologically and economically insignificant for 7 decades, has tracked very closely with the progress of the TACF breeding program (Fig. 4). In addition, much of the published research addresses questions directly related to the breeding program or implicitly assumes that breeding will solve the blight problem and permit the restoration of the species. We conservatively estimate that tens of millions of dollars from public and private sources has been invested since the 1980s in the work of TACF and complementary research at universities and in federal and state agencies, mostly in the expectation of a genetic solution to chestnut blight.
Number of published articles containing “American chestnut” or “Castanea dentata” in the title, by half-decade since 1970. Counted were articles on the biology or ecology of American chestnut, including breeding, transgenics, propagation, and silviculture. Excluded were false hits and articles dealing exclusively with the pathogen and disease.
The composite analyses from several years of progeny testing reported here have experimentally validated expectations that a blight-resistant population of American chestnut can be reconstructed using extra-specific alleles and backcross breeding. However, these analyses also demonstrate that ultimate success may require more generations of breeding than originally envisioned. This presents only minor technical difficulties compared with the administrative challenges of maintaining continuity in the breeding through philanthropy and grants. Near-term partial successes will be critical to sustaining enthusiasm. It is likely that both the breeding approach and the transgenic approach will play critical roles in the rescue of the species.
The rescue and restoration of American chestnut stands as one of the most difficult, prolonged, and complex single-species conservation tasks ever attempted. However, nine decades after chestnut breeding work began in the USDA and CAES programs, the reality of a solution is now less a matter of time and conjecture than has ever been the case before. Forest tree breeding is necessarily a difficult process, requiring a large amount of labor, many years of effort, and typically large amounts of land in isolated parcels. Incorporating new genes into the genome of a tree while preserving or recovering the essential character and genetic diversity of the species makes the task especially challenging.
American chestnut represents the rare case in which an introduced insect or pathogen represents an existential threat to a native species. In most well-known invasions the outcome so far has been that some fraction of the tree population escapes or is genetically resistant: e.g., white pine blister rust on species of Pinus subgenus Strobus, Dutch elm disease on European and American species of elm, beech bark disease on Fagus grandifolia Ehrh., emerald ash borer on American species of Fraxinus, Phytophthora lateralis on Chamaecyparis lawsoniana (A. Murr.) Parl., sudden oak death on Quercus agrifolia Née, butternut canker on Juglans cinerea L., and others (Sniezko et al. 2012). However, high single-species mortality rates can have cascading effects on ecosystem functioning (Stephenson 1986; Rizzo et al. 2005; Flower et al. 2013; Pautasso et al. 2013; Fieldler and McKinney 2014), and there is usually a significant conservation, if not economic, interest in assisting the recovery of such species through selection and breeding.
We tend to doubt that the TACF example of a species rescue can be duplicated for species that lack the compelling cultural stature and appeal of American chestnut. The fundamentally private character of the TACF model is too dependent upon philanthropy—and therefore personal appeal—to work with every species, and if there is neither an industrial champion of the species then government agencies must necessarily take the lead in such projects. However, we believe the success of TACF demonstrates the existence of a large and latent resource of “citizen scientists,” i.e., people willing to donate their labor or their land to participate in technical work alongside scientists in pursuit of a long-term conservation goal. Indeed, we have been continually amazed by the talent, enthusiasm, and dedication of TACF volunteers. Cultivating and coordinating this volunteer support requires staff time and thoughtful attention, but we submit that the potential benefit to long-term and complex tree breeding projects can be tremendous and perhaps even critical to success.
References
Anagnostakis SL (2012) Chestnut breeding in the United States for disease and insect resistance. Plant Dis 96:1392–1403
Baier KM (2010) Interspecific suppression subtractive hybridization identifies a differentially expressed Chinese chestnut (Castanea mollissima) laccase-like gene. M.S. thesis, State University of New York College of Environmental Science and Forestry, Syracuse, NY
Barakat A et al (2012) Chestnut resistance to the blight disease: insights from transcriptome analysis. BMC Plant Biol 12:38
Beavis WD (1994) The power and deceit of QTL experiments: Lessons from comparative QTL studies. In: DB Wilkinson (ed) 49th Annual corn and sorghum research conference American seed trade association, Chicago, IL, pp 250–266
Burnham CR (1981) Blight resistant American chestnut: there’s hope. Plant Dis 65:459–460
Burnham CR (1988) The restoration of the American chestnut. Am Sci 76:478–487
Burnham CR, Rutter PA, French DW (1986) Breeding blight-resistant chestnuts. Plant Breed Rev 4:347–397
Chen C, Sun Q, Narayanan B, Nuss DL, Herzberg O (2010) Structure of oxalacetate acetylhydrolase, a virulence factor of the chestnut blight fungus. J Biol Chem 285:26685–26696
Clapper RB (1952) Relative blight resistance of some chestnut species and hybrids. J For 50:453–455
Clark SL, Schlarbaum SE, Saxton AM, Hebard FV (2016) Establishment of American chestnuts (Castanea dentata) bred for blight (Cryphonectria parasitica) resistance: influence of breeding and nursery grading. N For 47:243–270
Crandall BS, Gravatt GF, Ryan MM (1945) Root disease of Castanea species and some coniferous and broadleaf nursery stocks, caused by Phytophthora cinnamomi. Phytopathology 35:162–180
D’Amico KM, Horton TR, Maynard CA, Stehman SV, Oakes AD, Powell WA (2014) Assessing ectomycorrhizal associations on transgenic American chestnut compared to the wild-type, a conventionally-bred hybrid, and related Fagaceae species. Appl Environ Microbiol 81:100–108
Dalgleish HJ, Nelson CD, Scrivani JA, Jacobs DF (2016) Consequences of shifts in abundance and distribution of American chestnut for restoration of a foundation forest tree. Forests 7(1):9
Diskin M, Steiner KC, Hebard FV (2006) Recovery of American chestnut characteristics following hybridization and backcross breeding to restore blight-ravaged Castanea dentata. For Ecol Manag 223:439–447
Donaldson PA, Anderson T, Lane BG, Davidson AL, Simmonds DH (2001) Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol Mol Plant Pathol 59:297–307
Dong X et al (2008) Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228:331–340
El-Kassaby YA, Lstiburek M (2009) Breeding without breeding. Genet Res 91:111–120
Fang G-C, Blackmon BP, Staton ME, Nelson CD, Kubisiak TL, Olukolu BA, Henry D, Zhebentyayeva T, Saski CA, Cheng C-H, Monsanto M, Ficklin S, Atkins M, Georgi LL, Barakat A, Wheeler N, Carlson JE, Sederoff R, Abbott AG (2012) A physical map of the Chinese chestnut (Castanea mollissima) genome, and its integration with the genetic map. Tree Genet Genomes 9:525–537
Fieldler CE, McKinney ST (2014) Forest structure, health, and mortality in two Rocky Mountain whitebark pine ecosystems: implications for restoration. Nat Areas J 34:290–299
Flower CE, Knight KS, Gonzalez-Meler MA (2013) Impacts of the emerald ash borer (Agrilus planipennis Fairmaire) induced ash (Fraxinus spp.) mortality on forest carbon cycling and successional dynamics in the eastern United States. Biol Invasions 15:931–944
Gingrich SF (1967) Measuring and evaluating stocking and stand density in uplant hardwood forests in the central states. For Sci 13:38–53
Grattapaglia D, Resende MDV (2011) Genomic selection in forest tree breeding. Tree Genet Genomes 7:241–255
Gravatt F (1949) Chestnut blight in Asia and North America. Unasylva 3:2–7
Graves AH (1950) Relative blight resistance in species and hybrids of Castanea. Phytopathology 40:1125–1131
Griffin, GJ, Elkins JR, McCurdy D, Griffin SL (2006) Integrated use of resistance, hypovirulence, and forest management to control blight in American chestnut. In: Steiner, KC and JE Carlson (eds) Restoration of American chestnut to forest lands, pp 97–107. Proceedings of a conference and workshop, May 2006, The North Carolina Arboretum. United States National Park Service Natural Resources Report NPS/NCR/CUE/NRR-2006/001
Griffin GJ, Hebard FV, Wendt RW, Elkins JR (1983) Survival of American chestnut trees: evaluation of blight resistance and hypovirulence in Endothia parasitica. Phytopathology 73:1084–1092
Havir EA, Anagnostakis SL (1983) Oxalate production by virulent but not by hypovirulent strains of Endothia parasitica. Physiol Plant Pathol 23:369–376
Havir EA, Anagnostakis SL (1985) Oxaloacetate acetylhydrolase activity in virulent and hypovirulent strains of Endothia (Cryphonectria) parasitica. Physiol Plant Pathol 26:1–9
He X, Miyasaka SC, Fitch MMM, Khuri S, Zhu YJ (2013) Taro (Colocasia esculenta) transformed with a wheat oxalate oxidase gene for improved resistance to taro pathogen Phytophthora colocasiae. HortScience 48:22–27
Hebard FV (1994) The American Chestnut Foundation breeding plan: beginning and intermediate steps. J Am Chestnut Found 8(1):21–28
Hebard FV (2002) Meadowview notes 2001–2002. J Am Chestnut Found 16(1):7–18
Hebard FV (2004) Research objectives of the American Chestnut Foundation, 2004–2014. J Am Chestnut Found 18(2):13–19
Hebard FV (2006) The backcross breeding program of The American Chestnut Foundation. In: Steiner KC, Carlson JE (eds) Restoration of American chestnut to forest lands, pp 61–77. Proceedings of a conference and workshop, May 2006, The North Carolina Arboretum. United States National Park Service Natural Resources Report NPS/NCR/CUE/NRR-2006/001
Hebard FV (2012) The American Chestnut Foundation breeding program. In: Sniezko RA et al (eds) Proceedings of the fourth international workshop on the genetics of host-parasite interactions in forestry: disease and insect resistance in forest trees, pp 221–234. USDA Forest Service General Technical Report PSW-GTR-240, 372 pp
Hebard FV, Shain L (1988) Effects of virulent and hypovirulent Endothia parasitica and their metabolites on ethylene production by bark of American and Chinese chestnut and scarlet oak. Phytopathology 78:841–845
Hebard FV, Griffin GJ, Elkins JR (1984) Developmental histopathology of cankers incited by virulent and hypovirulent Endothia parasitica on susceptible and resistant chestnut trees. Phytopathology 74:140–149
Hepting GH (1974) Death of the American chestnut. J For Hist 18:60–67
Hu X, Bidney DL, Yalpani N, Duvick JP, Crasta O, Folkerts O, Lu G (2003) Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol 133:170–181
Jacobs DF, Dalgleish HJ, Nelson CD (2013) A conceptual framework for restoration of threatened plants: the effective model of American chestnut (Castanea dentata) reintroduction. N Phytol 197:378–393
Jaynes RA (1978) Selecting and breeding blight-resistant chestnut trees. In: Proceedings of the American Chestnut symposium, Morgantown, WV, pp 4–6
Jeffers SN, Meadows IM, James JB, Sisco PH (2012) Resistance to Phytophthora cinnamomi among seedlings from backcross families of hybrid American chestnut. In: Sniezko RA et al (eds) Proceedings of the fourth international workshop on the genetics of host-parasite interactions in forestry: disease and insect resistance in forest trees, pp 194–195. USDA Forest Service Gen Tech Rep PSW-GTR-240, 372 pp
Jones C, Griffin GJ, Elkins JR (1980) Association of climatic stress with blight on Chinese chestnut in the eastern United States. Plant Dis 64:1001–1004
Kim KS, Min JY, Dickman MB (2008) Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol Plant Microbe Interact 21:605–612
Kubisiak TL, Hebard FV, Nelson CD, Jhang J, Bernatzky R, Huang H, Anagnostakis SL, Doudrick RL (1997) Molecular mapping of resistance to blight in an interspecific cross in the genus Castanea. Phytopathology 87:751–759
Kubisiak TL, Nelson CD, Staton ME, Zhebentyayeva T, Smith C, Olukolu BA, Fang G-C, Hebard FV, Anagnostakis S, Wheeler N, Sisco PH, Abbott AG, Sederoff RR (2013) A transcriptome-based genetic map of Chinese chestnut (Castanea mollissima) and identification of segmental homology with peach (Prunus persica). Tree Genet Genomes 9:557–571
Liang H, Maynard CA, Allen RD, Powell WA (2001) Increased Septoria musiva resistance in transgenic hybrid poplar leaves expressing a wheat oxalate oxidase gene. Plant Mol Biol 45:619–629
Livingstone DM, Hampton JL, Phipps PM, Grabau EA (2005) Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant Physiol 137:1354–1362
Lstiburek M, Hodge GR, Lachout P (2015) Uncovering genetic information from commercial forest plantations—making up for lost time using “breeding without breeding”. Tree Genet Genomes 11: Article 55
Maynard CA, Polin LD, LaPierre S, Rothrock RE, Powell WA (2006) American chestnut (Castanea dentata (Marsh.) Borkh.). In: Wang K (ed) Agrobacterium protocols, 2nd edn. Methods in molecular biology book series #344. Humana Press, Inc., Totowa, NJ, pp 239–251
McKay JW, Jaynes RA (1969) Chestnuts. In: Jaynes RA (ed) Handbook on North American nut trees. Northern Nut Growers’ Association, Knoxville, pp 264–286
Merkle SA, Wiecko AT, Watson-Pauley BA (1991) Somatic embryogenesis in American chestnut. Can J For Res 21:1698–1701
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Milgroom MG, Cortesi P (2004) Biological control of chestnut blight with hypovirulence: a critical analysis. Annu Rev Phytopathol 42:311–338
Namkoong G (1991) Maintaining genetic diversity in breeding for resistance in forest trees. Ann Rev Phytopathol 29:325–342
Nelson CD, Powell WA, Merkle SA, Carlson JE, Hebard FV, Islam-Faridi N, Staton ME, Georgi L (2014) Chestnut. In: Ramawat K (ed) Tree biotechnology, chapter 1. CRC Press, Boca Raton, pp 3–35
Newhouse JR (1990) Chestnut blight. Sci Am 262(1):106–111
Newhouse AE, McGuigan LD, Baier KA, Valletta KE, Rottmann WH, Tschaplinski TJ, Maynard CA, Powell WA (2014) Transgenic American chestnuts show enhanced blight resistance and transmit the trait to T1 progeny. Plant Sci 228:88–97
Nyland RD (1996) Silviculture concepts and applications. The McGraw-Hill Companies, New York, p 633
Paillet FL (2002) Chestnut: history and ecology of a transformed species. J Biogeogr 29:1517–1530
Partridge-Telenko DE, Hu J, Livingstone DM, Shew BB, Phipps PM, Grabau EA (2011) Sclerotinia blight resistance in Virginia-type peanut transformed with a barley oxalate oxidase gene. Phytopathology 101:786–793
Pautasso M, Aas G, Queloz V, Holdenrieder O (2013) European ash (Fraxinus excelsior) dieback—a conservation biology challenge. Biol Conserv 158:37–49
Polin LD, Liang H, Rothrock R, Nishii M, Diehl D, Newhouse A, Nairn CJ, Powell WA, Maynard CA (2006) Agrobacterium-mediated transformation of American chestnut (Castanea dentata (Marsh.) Borkh.) somatic embryos. Plant Cell Tissue Organ Cult 84:69–79
Rizzo DM, Garbelotto M, Hansen EM (2005) Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests. Ann Rev Phytopathol 43:309–335
Schneider M, Droz E, Malnoë P, Chatot C, Bonnel E, Métraux JP (2002) Transgenic potato plants expressing oxalate oxidase have increased resistance to oomycete and bacterial pathogens. Potato Res 45:177–185
Sisco PH, Kubisiak TL, Casasoli M, Barreneche T, Kremer A, Clark C, Sederoff RR, Hebard FV, Villani F (2005) An improved genetic map for Castanea mollissima/Castanea dentata and its relationship to the genetic map of Castanea sativa. Acta Hortic 693:491–496
Sniezko RA, Yanchuk AD, Kliejunas JT, Palmieri KM, Alexander JM, Frankel SJ (Technical Coordinators) (2012) Proceedings of the fourth international workshop on the genetics of host-parasite interactions in forestry: disease and insect resistance in forest trees. Gen Tech Rep PSW-GTR-240. Pacific Southwest Research Station, Forest Service, U.S. Department of Agriculture, Albany, CA, p 372
Staton ME, Zhebentyayeva T, Olukolu B, Fang GC, Nelson CD, Carlson JE, Abbott AG (2015) Substantial genome synteny preservation among woody angiosperm species: comparative genomics of Chinese chestnut (Castanea mollissima) and plant reference genomes. BMC Genomics 16: Article 744
Stephenson SL (1986) Changes in a former chestnut-dominated forest after a half century of succession. Am Midl Nat 116:173–179
Stephenson SL, Adams HS, Lipford ML (1991) The present distribution of chestnut in the upland forest communities of Virginia. Bull Torrey Bot Club 118:24–32
Tarcali G, Radocz L (2009) Experiences of a study trip in China on the research of chestnut blight fungus and gall wasp. Analele Universităţii din Oradea, Fascicula: Protecţia Mediului 14:410–419
Walz A, Zingen-Sell I, Loeffler M, Sauer M (2008) Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum. Plant Pathol 57:453–458
Wheeler NC, Steiner KC, Schlarbaum SE, Neale DB (2015) The evolution of forest genetic and tree improvement research in the United States. J For 113:500–510
Yan B, Li Z, Huang H, Qin L (2007) Genetic diversity and population differentiation of chestnut blight fungus. Biochem Genet 45:487–506
Zhang B, Oakes AD, Newhouse AE, Baier KM, Maynard CA, Powell WA (2013) A threshold level of oxalate oxidase transgene expression reduces Cryphonectria parasitica-induced necrosis in a transgenic American chestnut (Castanea dentata) leaf bioassay. Transgenic Res 22:973–982
Zhebentyayeva T et al (2013) Genetic and genomic resources for mapping resistance to Phytophthora cinnamomi in chestnut. Acta Hortic 1019:263–270
Author information
Authors and Affiliations
Corresponding author
Additional information
Frederick V. Hebard and Laura L. Georgi—Retired The American Chestnut Foundation.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Steiner, K.C., Westbrook, J.W., Hebard, F.V. et al. Rescue of American chestnut with extraspecific genes following its destruction by a naturalized pathogen. New Forests 48, 317–336 (2017). https://doi.org/10.1007/s11056-016-9561-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11056-016-9561-5