Abstract
Pythium oligandrum (Oomycota) is known for its strong mycoparasitism against more than 50 fungal and oomycete species. However, the ability of this oomycete to suppress and kill the causal agents of dermatophytoses is yet to be studied. We provide a complex study of the interactions between P. oligandrum and dermatophytes representing all species dominating in the developed countries. We assessed its biocidal potential by performing growth tests, on both solid and liquid cultivation media and by conducting a pilot clinical study. In addition, we studied the molecular background of mycoparasitism using expression profiles of genes responsible for the attack on the side of P. oligandrum and the stress response on the side of Microsporum canis. We showed that dermatophytes are efficiently suppressed or killed by P. oligandrum in the artificial conditions of cultivations media between 48 and 72 h after first contact. Significant intra- and interspecies variability was noted. Of the 69 patients included in the acute regimen study, symptoms were completely eliminated in 79% of the patients suffering from foot odour, hyperhidrosis disappeared in 67% of cases, clinical signs of dermatomycoses could no longer be observed in 83% of patients, and 15% of persons were relieved of symptoms of onychomycosis. Our investigations provide clear evidence that the oomycete is able to recognize and kill dermatophytes using recognition mechanisms that resemble those described in oomycetes attacking fungi infecting plants, albeit with some notable differences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dermatophytic fungi of the genera Trichophyton, Nannizzia, Microsporum and Epidermophyton cause infections of keratinized structures such as the skin, hairs and nails of healthy individuals [1]. Although infections by dermatophytes are usually not life-threatening, they are widespread and difficult to eliminate completely [2]. According to World Health Organization statistics, the global prevalence of dermatophytoses is approaching 20–25%, making it one of the most frequent infectious diseases, with treatment costs estimated at half a billion dollars annually [3, 4]. Dermatophytes are grouped ecologically according to their habitat as being either anthropophilic (human-associated), zoophilic (animal-associated) or geophilic (soil-dwelling). Virulence factors of dermatophytes remained unknown until recently, when comparative genome analyses revealed candidate genes possibly involved in the infection process [5,6,7]. Three different classes of genes and their products are cited most often as critical factors: proteases secreted to degrade skin, kinases involved in signalling necessary for the interaction between the host and the fungus and LysM adhesins that appear to bind to surface carbohydrates of dermatophytes and mask them from the immune response of the host. These latter factors, in particular, appear to be responsible for the poor recognition of dermatophytes by keratinocytes and macrophages, and the subsequent inhibition of an effective immune response of infected organisms against dermatophytes.
Considering the low efficiency of chemical antifungals against dermatophytes [8, 9], their elimination using biological defence means would appear as an attractive alternative. However, the biological enemy will have to offer universal and safe elimination mechanisms, considering the physiological and etiological variability among individual dermatophytes. Pythium oligandrum is a non-pathogenic soil-inhabiting peronosporomycete (oomycete) colonizing the root ecosystems of many crop species [10]. This microorganism exhibits strong mycoparasitism against more than 50 fungal and oomycete species, including some of its relatives [11]. To become such an efficient parasite, P. oligandrum has developed a number of traits allowing it to recognize, engage and destroy target fungi [12, 13] and it is assumed that it acquired the parasitism genes from the three eukaryotic kingdoms and from bacteria [14]. Hydrolytic enzymes, such as chitinases, cellulases, proteases and glucanases, secreted by P. oligandrum are often cited as molecular tools critical for their mycoparasitic success [10, 15]. Competition for space and nutrients are other mechanisms used by P. oligandrum for biological control [16]. The unique possibilities of this microorganism have been used extensively for the protection of plants from fungi [11, 16,17,18,19]. In addition, P. oligandrum has found practical use in human and veterinary medicine for the elimination of dermatophytes [19, 20]. Despite the practical medical observations cited above, the exact cellular and molecular mechanisms behind the elimination of the causal agents of dermatophytoses have not been investigated.
Here we provide a systemic study of the interactions between P. oligandrum and dermatophytes representing all ecological groups and species that dominate in developed countries [21, 22]. We assessed its biocidal potential by performing growth tests, both on solid and on liquid cultivation media, and by conducting a clinical study. We studied the molecular background of mycoparasitism using expression profiles of genes responsible for the attack on the side of P. oligandrum and the stress response on the side of Microsporum canis. Our investigations provide clear evidence that the oomycete is able to recognize and kill dermatophytes using recognition mechanisms that resemble those described in oomycetes attacking fungi infecting plants, albeit with some notable differences.
Materials and Methods
Microbial Strains and Media
The M1 strain of P. oligandrum was provided by the company Biopreparáty, Ltd (Únětice, Czech Republic) and corresponds to strain ATCC 38472. This oomycete was isolated from sugar beet [18, 23]. Ten different species and 23 different strains of dermatophytes were obtained from the CCF collection (Culture Collection of Fungi, Charles University, Prague, Czech R.), from the CCM collection (Czech Collection of Microorganisms, Masaryk University, Brno, Czech R.) or from the working collection kept at the Institute of Microbiology of the Czech Academy of Sciences. Competition tests on solid media were carried out for the most common species of dermatophytes, including one strain of Epidermophyton floccosum, one strain of M. canis, two strains of Nannizzia fulva (syn. Microsporum fulvum), two strains of N. gypsea (Microsporum gypseum), two strains of N. persicolor (syn. Microsporum persicolor), five strains of Trichophyton benhamiae (syn. Arthroderma benhamiae), one strain of Trichophyton erinacei, four strains of Trichophyton interdigitale, three strains of Trichophyton rubrum and one strain of Trichophyton tonsurans (Online Resource 1). The strain M. canis CCM 8353 was used for the gene expression study. The identity of all strains was verified using ITS rDNA barcode sequence and PCR fingerprinting comparisons with reference strain according to Hubka et al. [24].
Interaction Studies on Plates
The interaction between the dermatophytes and P. oligandrum was done on malt extract agar (MEA, malt extract, 20 g/l, d-glucose, 20 g/l, peptone, 1 g/l, agar 20 g/l) and potato dextrose agar (PDA, HI Media) incubated at 25 °C in the dark. First, the dermatophytes under examination were inoculated on one side of the plate and allowed to grow for 3–10 days, producing colonies 20–25 mm in size. Thereafter, the agar block with P. oligandrum was placed onto the opposite side of the Petri dish, and the continuation of the growth of the dermatophyte and Pythium were evaluated every 2 days until 10 days. The measured parameter was the percentage occupancy of plates calculated as the distance of the front of the growing microorganism from the edge of the Petri dish (in mm) divided by the diameter of the plate (80 mm) and multiplied by 100. Each experiment was performed in triplicate on each of the media used. At the end of the experiment, the viability of the tested dermatophytes was also evaluated. An agar block (1 × 1 cm) was cut from the interaction zone, where both organisms were visibly present, and transferred to Czapek-Dox agar (CDA, sucrose 30 g/l, agar 20 g/l, NaN03 3 g/l, K2HPO4 1 g/l, MgSO4 0.5 g/l, KCl, 0.5 g/l, Fe2(SO4)3 0.01 g/l, pH 6.5), which enables the growth of the dermatophytes but not of P. oligandrum.
Interaction Studies in Suspensions
The evaluation of the P. oligandrum fungicidal effect was according to EU Standards [ČSN EN 1275: 2006: Chemical disinfectants and antiseptics—quantitative suspension test for the evaluation of basic fungicidal or basic yeasticidal activity of chemical disinfectants and antiseptics—test method and requirements (phase 1)] and was conducted by the test laboratory Chemila, spol. s r.o. (Hodonín, Czech R., accredited as the test laboratory No. 1273 by Czech Accreditation Institute according to the norm ČSN EN ISO/IEC 17025). The strains used were Trichophyton rubrum strain 584/2017, Trichophyton interdigitale CCM 8377 and M. canis CCM 8353 (Online Resource 1). Test suspensions of dermatophytes were prepared by washing the spores with 0.05% polysorbate 80 in water, gentle shaking with the glass beads and filtration through a fritted filter with porosity 40–100 μm. The tested preparation of P. oligandrum (batch No. 060217.3, BARD s.r.o.) was resuspended in the distilled water to get 1% suspension with concentration ranging from 100 to 200 CFU/ml and activated for 30 min at 20 °C. During the suspension test, 0.5 ml of dermatophyte spore suspensions (density of 3.04 × 106/ml for T. rubrum, 0.88 × 106/ml for T. interdigitale and 0.26 × 106/ml for M. canis) was mixed with 0.5 ml of the P. oligandrum suspension and incubated at 20 ± 1 °C for 1 h, 24 h and 48 h. Subsequently, tenfold serial dilutions were prepared from the incubated suspensions and number of CFU was evaluated by the cultivation on the Sabouraud agar for 7–10 days at 25 °C. Results are expressed as logarithmic microbial viability reduction for each test microorganism designated as log R (reduction of vitality). The log R is calculated based on the formula log R = log N0 − log NE, which accounts the concentration (CFU/ml) of the dermatophyte at the beginning (N0) and at the end of the contact time (NE) with P. oligandrum.
Gene Expression Profiles on Agar Plates and in the Liquid Suspension
The three genes connected with mycoparasitism on the side of P. oligandrum [10, 15] and aggressivity and stress response on the side of dermatophyte [6] were selected for the understanding of the molecular mechanisms standing behind interaction of both organisms (Table 1). For gene expression profiles on agar plates, samples of both the dermatophyte and P. oligandrum were taken by cutting the agar blocks (4 × 8 mm) from the interaction zone and zone with pure P. oligandrum or dermatophyte as is shown in Fig. 4. The lower part of the block containing the agar was removed, and the upper part with the mycelium was used for DNA and RNA extraction using the protocol of Berendzen et al. [25]. Gene expression profiling started from day 4 (just before the physical contact of both microbes) and proceeded until day 7. For gene expression profiles in liquid suspension, conidia of M. canis CCM 8353 were prepared and counted using the method described by Saunte et al. [26]. A suspension of P. oligandrum was obtained using a liquid culture and counted on the basis of all reproductive forms, sporangia, zoospores and oospores [23]. To initiate the interaction experiment, 4 ml of MEA (without agar) medium was mixed with 0.5 ml of M. canis conidia (5 × 106 conidia/ml) and 0.5 ml of P. oligandrum (as the sum of reproductive forms, 5 × 106 cells/ml) in six-well plastic plates (BioTech, Czech Republic) and incubated at 30 °C. In control experiments, one of the microorganisms was omitted and replaced by a pure medium. From each well, 50 µl of the liquid medium was collected in 6-h intervals for subsequent culturing on CDA plates for viability testing and gene expression profiling. Good aeration and homogeneity of the sample were assured by its frequent agitation.
The primers were used based on the cited literature or designed with the Primer-Blast tool (http://ncbi.nlm.nih.gov/tools/primer-blast) (Table 1). Specificity of the PCR primers was confirmed by the sequencing of their PCR products. All DNA/RNA amplifications were performed using the CFX Connect Real-Time PCR Detection System operated using the CFX Manager™ Software ver. 3.0 (BioRad) as described previously [27]. Briefly, reverse transcription reactions were performed in Hard-Shell® 96-Well PCR plates (BioRad) in the total reaction volume of 10 µl composed of 5 µl of the individual, stabilized RNA samples and 5 µl of the reverse transcription master mix A + B (Generi Biotech, Czech Republic). Each plate was sealed and incubated at 42 °C for 60 min. Thereafter, the reaction mixtures were diluted 50× using RNAse-free water according to the instructions of the manufacturer. The qPCR amplifications were performed in Hard-Shell® 96-Well PCR plates in the total reaction volume of 10 µl composed of 4 µl of diluted cDNA (see above), 1 µl of the primer mixture containing 5 µM of each of the corresponding forward and reverse primer, and 5 µl of SsoAdvanced Universal SYBR® Green Supermix (BioRad). The plates were placed into the CFX Connect Real-Time PCR System (BioRad) operated using CFX Manager™ Software. The cycling parameters were: 95 °C for 3 min followed by 50 cycles of 95 °C for 10 s and 60 °C for 30 s. The data were evaluated using the 2−ΔΔCt (Livak) method as described in the Real-Time PCR Application Guide (BioRad). The expression of inducible genes was related to the expression of two constitutive reference genes for beta tubulin and glyceraldehyde-3-phosphate dehydrogenase, which expression varies within the range ± 50% under the range of tested conditions [28, 29]. Gene expression on the interaction plate was corrected based on gene expression on the control plate containing the single microorganisms under examination [27].
Clinical Study
This study is a retrospective clinical trial conducted at the Department of Dermatology and Venerology of the Pardubice Regional Hospital in Pardubice between 1 June 2007 and 1 June 2014. No randomization of patients was performed. Informed consent was obtained from all individual participants included in the study. Patients were mostly outpatients included in the study based on the following criteria. For the study of acute patients (n = 69), these criteria were: clinical symptoms of foot mycoses confirmed by either microscopic observation or a positive microbial cultivation test. For the recurrent infection study (n = 29), the criteria were: recurring problems with tinea interdigitalis infection at least twice yearly, an acute attack of tinea interdigitalis present upon entering the study and confirmation of foot mycosis by both microscopy and cultivation. No patients had to be excluded from the study on the basis of their age or clinical status. From the set of 29 patients with recurrent infection, 16 had an acute attack of tinea interdigitalis at least twice yearly, 9 of them three times a year, 3 patients stated four attacks annually, and 1 person suffered problems with the disease continuously. In acute patients, the profile of the causative agents revealed using cultivation was consistent with published data, with T. rubrum and T. interdigitale (8) representing two of the by far most common species (Online Resource 2). In patients with recurrent infection, the most common causal agent was T. rubrum (72% of patients), sometimes in combination with candidiasis. Pythium oligandrum was applied in the form of the cosmetic product Biodeur ® (Bio Agens Research and Development, BARD s.r.o., Únětice, Czech R.) which is composed of P. oligandrum M1 dried spores (> 2 × 105 oospores/g) stored in the presence of a silica desiccative and dried millet (Panicum miliaceum), which provides the substrate for the revitalization of the microorganism. For the foot baths, 1 g of the Biodeur ® was reconstituted in 2–3 l of tap water warmed to 34 °C. Patients’ feet were washed in this solution for 30 min and allowed to dry spontaneously. For acute infections, the foot bath was applied on two alternate days in three consecutive weeks, and the medical evaluation was performed 1 month after the last bath. The protocol for recurrent tinea interdigitalis patients was based on the initial bolus, identical to the acute situation with additional applications in weeks 5, 7 and 10, followed by maintenance applications 6 weeks apart. As an additional preventive measure, shoe spraying by Biodeur ® suspension was applied twice per week in weeks 1–5, followed by once per week sprays in weeks 6–9, and additional sprays every second week thereafter. In the study of patients with recurrent tinea interdigitalis, continuous monitoring in the form of regular checks followed, the time of the cessation of the disease was evaluated based on clinical evaluation supplemented by mycological evaluation by microscopy and cultivation.
Results
Patterns and Kinetics of Interaction Between P. oligandrum and Dermatophytes on Plates and in the Suspension
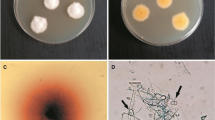
The three types of growth pattern scenarios were observed. Firstly, the growth of P. oligandrum over the dermatophyte was observed in case of M. canis, N. persicolor, T. benhamiae, T. rubrum and T. tonsurans (Fig. 1). Concerning the kinetics of the interaction, T. benhamiae, T. rubrum or T. tonsurans, got rapidly overgrown by the oomycete. In M. canis, the curve is biphasic, indicating a certain degree adaptation to Pythium in the initial stage (Fig. 2). In other species, the interaction started with the formation of a contact inhibition zone of various intensity, varying from wide (N. fulva, T. erinacei, T. interdigitale and E. floccosum) to narrow (N. gypsea) (Fig. 1). This interaction occurred at the level of substrate mycelium and was later followed by the production of aerial mycelium by P. oligandrum, which overgrew the dermatophyte species. The last category of dermatophytes includes N. fulva and some strains of N. gypsea, N. persicolor and T. interdigitale that were able to adapt to the attack mounted by Pythium and, eventually, to stop its action.
Examples of the time course of direct interactions representing all five interaction patterns. Type I—exponential single phase of the ascending type. a Trichophyton rubrum CCF 4933. b Trichophyton benhamiae CCF 4918. Type II—exponential two-phase pattern of the ascending type. c Trichophyton erinacei CCF 4472. Type III—exponential single phase with an ascending and a descending phase. d. Nannizzia persicolor CCF 4542. e Epidermophyton floccosum PL 231. Type IV—two-phase pattern with an ascending and a descending phase. f. Nannizzia gypsea CCF 4626. The photograph was taken on days 4, 6, 8 and 10 of the experiment, ordered sequentially from left to right
Examples of the time course of the elimination of dermatophytes (gray line) by Pythium oligandrum (black line) on the MEA cultivation medium. a Epidermophyton floccosum. b Microsporum canis. c Nannizzia fulva d Nannizzia gypsea. e Nannizzia persicolor. f Trichophyton benhamiae. g Trichophyton erinacei. h Trichophyton interdigitale. i Trichophyton rubrum. j Trichophyton tonsurans. Error bars represent the standard deviation counted from all strains and Petri dishes of the particular dermatophyte species
The competitive ability of P. oligandrum on MEA was generally better than on PDA, with the exception of N. fulva and certain strains of T. interdigitale. Excluding species from which only a single strain was available, notable intraspecies variability was found in N. fulva, N. gypsea and N. persicolor (Fig. 3).
Viability tests were conducted after the dermatophyte overgrown by P. oligandrum was transferred to CDA medium allowing the growth of the dermatophyte, but not of the Pythium. Using this approach, it could be shown that the dermatophytes were actually dead after their interaction with the Pythium.
Under the conditions of the liquid culture, the 3–4 log viability reduction of the target dermatophyte after 48 h was observed (Table 2).
Gene Expression Profiles and Viability During the Elimination of Microsporum canis on the Plate
The results regarding the expression of inducible aggressivity genes in P. oligandrum and M. canis are detailed in Fig. 4. Only expression relative to beta tubulin is shown since the expression relative to glyceraldehyde-3-phosphate dehydrogenase provided very similar data (not shown). Concerning the oomycete, on day 4 the expression of all genes was very low compared to the control situation. After the direct contact on day 5, the genes for the critical digestion enzyme cellulose (POCELL) got switched on, followed by the expression of cell wall lysing endo-β-1,3-glucanase (POENDO) and sporulation marker POSTRU with the peak on the day 6. On the day 7, only the POSTRU expression persisted, when the oomycete had most probably exhausted the available nutrients and switched its metabolism to the sporulation mode.
Gene expression profiles during the interaction of Pythium oligandrum with the dermatophyte Microsporum canis. An agar block with Pythium oligandrum was added on day 3 onto a Petri dish with a well-grown dermatophyte. Photodocumentation and gene expression profiling started from day 4 and proceeded until day 7. For Pythium oligandrum, we examined the expression of genes coding for cellulase (POCELL), endo-β-1,3-glucanase (POENDO) and the tyrosine-rich structural protein (POSTRU), whereas for Microsporum canis we followed the expression of genes for the LysM adhesion/masking protein (MCLYSM), metalloproteinase (MCMETA) and Ca-dependent kinase (MCCAMK). The results are shown at logarithmic scale; each bar shows the average value for three independent experiments
In case of dermatophyte, there was considerable upregulation of all genes before the contact of both taxa on the day 4 and after the contact on the day 5. Then, on day 6, the biological antagonism between the oomycete and the dermatophytes was decided for the oomycete. This was documented by the dramatic cessation of the expression of all analysed genes, with only negligible amounts of transcripts being detected.
Gene Expression Profiles and Viability During the Elimination of Microsporum canis Conidia in Suspension
The gene expression profiles under the suspension conditions showed the same pattern as observed in the plate experiment (Figs. 4, 5). The notable difference was in the expression of the POENDO gene which preceded the expression of the POCELL gene, and in the normalized expression levels were generally much lower. Furthermore, while the dermatophyte retained considerable overexpression of the MCLYSM and MCMETA genes, we observed very little expression of the MCCAMK gene. The cultivation experiment shows that the development of the dermatophyte could be suppressed completely within 60 h (Online Resource 3).
Gene expression profiles and viability during the interaction of Pythium oligandrum with the dermatophyte Microsporum canis in suspension. For Pythium oligandrum, we examined the expression of genes coding for cellulase (POCELL), endo-β-1,3-glucanase (POENDO) and the tyrosine-rich structural protein (POSTRU) whereas while for Microsporum canis, we followed the expression of genes for the LysM effector/masking protein (MCLYSM), metalloproteinase (MCMETA) and Ca-dependent kinase (MCCAMK). The results are shown at logarithmic scale; each bar shows the averaged value for three independent experiments
Clinical Efficacy of Pythium oligandrum in Patients with Tinea Pedis
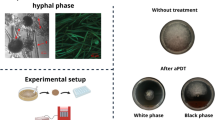
The efficacy of relief of foot mycoses symptoms is summarized in Fig. 6. Of the 69 patients included in the acute regimen study, 42 had odour symptoms, 43 exhibited hyperhidrosis, 58 had dermatomycosis, and 59 suffered from onychomycosis. Symptoms were completely eliminated in 79% of the patients suffering from foot odour, hyperhidrosis disappeared in 67% of cases, clinical signs of dermatomycoses could no longer be observed in 83% of patients, and 15% of persons were relieved of symptoms of onychomycosis (Fig. 6b). In patients with recurring infections, in 28 (97%) of all 29 patients, clinical symptoms disappeared within 6 weeks after the first application. Concerning the long-term follow-up of all 29 patients, only three patients that finished the 12-month application protocol had a single further episode of tinea interdigitalis within the next 3 months. The application of the biological cosmetic product containing P. oligandrum was well tolerated. We did not observe a single episode of an allergic reaction or any other side effect in either the acute or the recurrent group of patients (Fig. 6c).
Discussion
The ability of P. oligandrum to parasite on other fungi and oomycetes has been known since the forties of the last century [30]. However, this important aspect of the biology of this peronosporomycete has so far been analysed only in relation to its ability to provide protection against plant pathogens (reviewed in [10, 31]). We used three approaches to perform our investigations: direct observations of interactions in dual cultures, molecular analyses of gene expression profiles of both interacting microorganism and clinical tests in real conditions. In the cases of dermatophytes investigated on the Petri dishes, we could distinguish at least four patterns of interaction (Fig. 2): pattern I, defined as an exponential single phase of the ascending type (T. benhamiae, T. rubrum and T. tonsurans); pattern II, defined as an exponential two-phase pattern of the ascending type (M. canis and T. erinacei); pattern III, defined as an exponential single phase with an ascending and a descending phase (E. floccosum N. fulva and N. persicolor); and, lastly, pattern IV, defined as a two-phase pattern with an ascending and a descending phase (e.g. N. gypsea and T. interdigitale). It may be assumed that such patterns might reflect features of the more detailed mechanisms of mutual interactions controlled by consecutive waves of mutual recognition of the target dermatophyte by the oomycete, or consecutive waves of diffusion of soluble molecular factors (the detailed nature of which is yet to be ascertained).
In general, the results of the biological fight between the oomycete P. oligandrum and the target fungus is expected to be both species- and medium- (environment-) specific, and the presented observations confirm this (Fig. 3). Nevertheless, the entire group, including T. rubrum, a dominant dermatophyte species, is mostly killed and eliminated by P. oligandrum with high efficiency. The high efficiency of the biological elimination of the target dermatophyte species is further corroborated by the suspension interaction results, where the log suppression reached a 3–4 log reduction within 48 h (Table 2) that is required for chemical biocides [32]. Such uniformly high efficiency of killing is not common in other groups of fungi, where more extensive variations between the efficiency of killing individual fungi are observed [30].
Our molecular analyses provided an additional understanding of the molecular nature of the antagonism between the oomycete and the dermatophytes. It was interesting to observe that while in the case of P. oligandrum direct contact with its prey appeared to initiate very little changes in the gene expression profiles, the dermatophytes must have been informed about the presence of the biological enemy long before the actual contact (dramatic gene upregulation on day 4, Fig. 4). We hypothesize that volatile or diffusible compounds produced by P. oligandrum (reviewed in [16]) can mediate this kind of early response in the attacked fungus.
In case of the gene expression profiles of P. oligandrum, the sequence of gene upregulation could be explained based on our knowledge of the biological functions of the individual gene markers. Both cellulase (POCELL) and endo-β-1,3-glucanase (POENDO) belong to the group of glycohydrolase, which are able to digest the cell wall of the attacked fungus [33, 34]. However, remodelling of the own cell wall of the oomycete allows the endoglucanase to feature also as an important marker of sporulation [35]. The tyrosine-rich structural protein (POSTRU) has recently been identified as one of the most specific markers of sporulation and is thus expressed during the late stages [36]. Indeed, the sequence of gene expression in the plate experiment started with the upregulation of cellulase on day 5 of the experiment, especially in the samples taken from proximal areas of the contact zone between the oomycete and the dermatophyte. All three followed genes of the oomycete were significantly upregulated on day 6 of the experiment, albeit the cellulase only in the contact areas (Fig. 4). On day 7 of the experiment, sporulation of the oomycete was expected, which was supported by the persistent upregulation of the tyrosine-rich structural protein (Fig. 4). The time sequence of gene expression of the oomycete during the suspension experiment was somewhat different, reflecting different cellular populations and a different environment (Fig. 5). Here, the endoglucanase gene was expressed earlier compared to the plate experiments at 12 h after mixing, while both cellulase and endoglucanase genes are highly expressed at 24 h after mixing. The tyrosine-rich structural protein is expressed only at 48 h after mixing, when the sporulation phase occurred under the solution experimental conditions (Fig. 5).
The three dermatophyte genes followed in the present study are typical aggressivity and stress related genes [6]. LysM cell surface effector (MCLYSM) represents an important surface effector and masking protein that covers the surface of dermatophytes to block access of soluble protein recognition factors of the immune system. This mechanism is acting in plant-pathogenic fungi and in the response of dermatophytes to the human immune system [6]. We have shown that dermatophytes use this stress-induced pathway also in their response to mycoparasitism. Metalloprotease (MCMETA) is the principal digestion enzyme that allows dermatophytes to digest the skin protein keratin and thus mediates both the attachment of fungi to skin structures and their nutrition. Ca-dependent kinase (MCCAMK) is one of the most important signalling enzymes triggering and orchestrating the response of the dermatophytes. These genes were used because the genes that are upregulated during the biological struggle of the dermatophytes with the oomycete were not known. They were all switched very early on, showing that the dermatophytes were able to sense the enemy and react, and remained switched on until the moment of the death (Fig. 4). We again noticed some principal differences between the plate and the suspension experiment. In the plate experiment, the upregulation was much more dramatic and concerned all three followed genes on day 4 and day 5 of the experiment. In suspension, only the MCLYSM gene was upregulated at 12 h and accompanied by MCMETA at 24 h (Fig. 5).
In 2002, Mencl described the use of the cosmetic biopreparation Biodeur, containing a fermented millet substrate with a surface growth of P. oligandrum for the suppression of the hidrotic feet syndrome (foot sweating) and interdigital mycoses [19]. This author reported an effect 1 month after its application on patients having infections with T. rubrum, T. interdigitale, and other dermatophytes and yeasts. In patients subjected to that study, there was 78.6% elimination of odour symptoms, 67.4% elimination of the hidrotic symptom and 82.8% elimination of the dermatophyte infection, evidenced by the absence of the dermatophyte upon the microbial cultivation.
Our clinical results from the initial study can be viewed as remarkable, considering that dermatophytoses in humans are notoriously difficult to treat, and often recurring. We noticed a high efficiency of the cosmetic product containing oospores of the oomycete P. oligandrum in the elimination of clinical signs of dermatophytosis, odour symptoms and hyperhidrosis (83–67% of patients, n = 69). Onychomycoses are known to take much longer to resolve compared to other symptoms. Indeed, although notable recovery from interdigital damage could be observed as soon as 20 days after the first application of the biological products, as long as 9 months was needed to observe definitive signs of recovery in cases of onychomycosis (not shown).
In conclusion, the present study demonstrates the ability of the oomycete P. oligandrum to suppress or eliminate dermatophytes, emphasizing its efficiency against a broad spectrum encompassing virtually all clinically important species and demonstrating a susceptibility profile that has not been observed for the classes or species of fungi targeted by this oomycete. By using of viable P. oligandrum propagules, it was possible to prove that their suppressing effect on dermatophytes starts within hours of the mutual encounter and interaction. Our detailed description of such an aggressive type of parasitism provides a scope for the practical use of the findings presented here.
References
Weitzman I, Summerbell RC. The dermatophytes. Clin Microbiol Rev. 1995;8:240–59.
Achterman RR, White TC. A foot in the door for dermatophyte research. PLoS Pathog. 2012;8:e1002564. https://doi.org/10.1371/journal.ppat.1002564.
Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008;51:2–15. https://doi.org/10.1111/j.1439-0507.2008.01606.x.
Drake LA, Dinehart SM, Farmer ER, Goltz RW, Graham GF, Hordinsky MK, et al. Guidelines of care for superficial mycotic infections of the skin: tinea corporis, tinea cruris, tinea faciei, tinea manuum, and tinea pedis. J Am Acad Dermatol. 1996;34:282–6.
Achterman RR, White TC. Dermatophyte virulence factors: identifying and analyzing genes that may contribute to chronic or acute skin infections. Int J Microbiol. 2012;2012:358305. https://doi.org/10.1155/2012/358305.
Martinez DA, Oliver BG, Gräser Y, Goldberg JM, Li W, Martinez-Rossi NM, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. MBio. 2012;3:e00259-12. https://doi.org/10.1128/mBio.00259-12.
Chinnapun D. Virulence factors involved in pathogenicity of dermatophytes. Walailak J Sci Technol. 2015;12:573–80. https://doi.org/10.14456/WJST.2015.72.
Asz-Sigall D, Tosti A, Arenas R. Tinea unguium: diagnosis and treatment in practice. Mycopathologia. 2017;182:95–100. https://doi.org/10.1007/s11046-016-0078-4.
Kumar P, Latka C, Taneja B. Current antifungal therapy and drug resistance mechanisms in dermatophytes. In: Arora G, Sajid A, Kalia V, editors. Drug resistance in bacteria, fungi, malaria, and cancer. Springer: Cham; 2017. p. 371–85.
Benhamou N, le Floch G, Vallance J, Gerbore J, Grizard D, Rey P. Pythium oligandrum: an example of opportunistic success. Microbiology. 2012;158:2679–94. https://doi.org/10.1099/mic.0.061457-0.
Brožová J. Exploitation of the mycoparasitic fungus Pythium oligandrum in plant protection. Plant Prot Sci. 2002;38:29–35.
Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010;11:R73. https://doi.org/10.1186/gb-2010-11-7-r73.
Horner NR, Grenville-Briggs LJ, Van West P. The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation. Fungal Biol. 2012;116:24–41. https://doi.org/10.1016/j.funbio.2011.09.004.
Jiang RH, Tyler BM. Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol. 2012;50:295–318. https://doi.org/10.1146/annurev-phyto-081211-172912.
Picard K, Tirilly Y, Benhamou N. Cytological effects of cellulases in the parasitism of Phytophthora parasitica by Pythium oligandrum. Appl Environ Microbiol. 2000;66:4305–14. https://doi.org/10.1128/AEM.66.10.4305-4314.2000.
Gerbore J, Benhamou N, Vallance J, Le Floch G, Grizard D, Regnault-Roger C, et al. Biological control of plant pathogens: advantages and limitations seen through the case study of Pythium oligandrum. Environ Sci Pollut Res. 2014;21:4847–60. https://doi.org/10.1007/s11356-013-1807-6.
Deacon J. Studies on Pythium oligandrum, an aggressive parasite of other fungi. Trans Br Mycol Soc. 1976;66:383–91. https://doi.org/10.1016/S0007-1536(76)80206-9.
Veselý D. Studies of the mycoparasitism in rhizosphere of emerging sugar-beet. Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene Zweite Naturwissenschaftliche Abteilung: Mikrobiologie der Landwirtschaft, der Technologie und des Umweltschutzes. 1978;133:195–200.
Mencl K. Is it possible to influence dermatomycoses by ecological means? Cesk Dermatol. 2002;77:71–5.
Načeradská M, Mencl K. Biopreparation with Pythium oligandrum at the therapy of dermatophytosis in cats-case report. Veterinarstvi. 2010;60:559–63.
Seebacher C, Bouchara JP, Mignon B. Updates on the epidemiology of dermatophyte infections. Mycopathologia. 2008;166:335–52. https://doi.org/10.1007/s11046-008-9100-9.
Hubka V, Větrovský T, Dobiášová S, Skořepová M, Lysková P, Mencl K, et al. Molecular epidemiology of dermatophytoses in the Czech republic—two-year-study results. Cesko Slov Dermatol. 2015;89:167–74.
Martin FM. Inventor Biocontrol of fungal soilborne pathogens by Pythium oligandrum patent US Patent 5,961,971. 1999.
Hubka V, Dobiasova S, Dobias R, Kolarik M. Microsporum aenigmaticum sp. nov. from M. gypseum complex, isolated as a cause of tinea corporis. Med Mycol. 2014;52:387–96. https://doi.org/10.1093/Mmy/Myt033.
Berendzen K, Searle I, Ravenscroft D, Koncz C, Batschauer A, Coupland G, et al. A rapid and versatile combined DNA/RNA extraction protocol and its application to the analysis of a novel DNA marker set polymorphic between Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. Plant Methods. 2005;1:4. https://doi.org/10.1186/1746-4811-1-4.
Saunte D, Simmel F, Frimodt-Moller N, Stolle L, Svejgaard E, Haedersdal M, et al. In vivo efficacy and pharmacokinetics of voriconazole in an animal model of dermatophytosis. Antimicrob Agents Chemother. 2007;51:3317–21. https://doi.org/10.1128/AAC.01185-06.
Klimeš R, Suchánek M, Maštalková L, Gopfert E, Nedbalcová K, Blahutová A et al. Comparison of the efficacy of treatment of dermatophytosis in guinea pigs by chemical vs. biological antifungals: ecological peronosporomycete Pythium oligandrum is as efficient as the chemical antifungal enilconazole. Vet Dermatol. 2018. (in press).
Zhu Y, Zhou P, Hu J, Zhang R, Ren L, Li M, Ning F, Chen W, Yu L. Characterization of Pythium transcriptome and gene expression analysis at different stages of fermentation. PLoS ONE. 2013;8:e65552. https://doi.org/10.1371/journal.pone.0065552.
Kozera B, Raoacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54:391–406. https://doi.org/10.1007/s13353-013-0173-x.
Lutchmeah R, Cooke R. Aspects of antagonism by the mycoparasite Pythium oligandrum. Trans Br Mycol Soc. 1984;83:696–700.
Whipps JM. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot. 2001;52:487–511.
Barros MES, Santos DA, Hamdan JS. In vitro methods for antifungal susceptibility testing of Trichophyton spp. Mycol Res. 2006;110:1355–60. https://doi.org/10.1016/j.mycres.2006.08.006.
Campion C, Massiot P, Rouxel F. Aggressiveness and production of cell-wall degrading enzymes by Pythium violae, Pythium sulcatum and Pythium ultimum, responsible for cavity spot on carrots. Eur J Plant Pathol. 1997;103:725–35.
Natarajan C, Gupta V, Kumar K, Prasanna R. Molecular characterization of a fungicidal endoglucanase from the cyanobacterium Calothrix elenkinii. Biochem Genet. 2013;51:766–79. https://doi.org/10.1007/s10528-013-9605-x.
Judelson HS. The genetics and biology of Phytophthora infestans: modern approaches to a historical challenge. Fungal Genet Biol. 1997;22:65–76.
Grenville-Briggs LJ, Horner NR, Phillips AJ, Beakes GW, Van West P. A family of small tyrosine rich proteins is essential for oogonial and oospore cell wall development of the mycoparasitic oomycete Pythium oligandrum. Fungal Biol. 2013;117:163–72. https://doi.org/10.1016/j.funbio.2013.01.001.
Acknowledgements
The authors acknowledge the excellent technical assistance of Tomáš Vaněk and Adam Štípek from the QC Laboratories of Biopreparaty in Černčice. Alena Gabrielová, V. Hubka and M. Kolařík were supported by the Ministry of Health of the Czech Republic, grant No. AZV 17-31269A. Research and development of new biocides at BARD is supported by TRIO project FV20630 from the Ministry of Industries and Trade of Czech Republic.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Martin Suchánek and Radim Klimeš are owners and stakeholders in the companies Biopreparáty and Bio Agens Research and Development manufacturing biological antifungal products based on Pythium oligandrum.
Research Involving Human Participants and/or Animals
For this type of study formal consent is not required.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Handling Editor: Vishnu Chaturvedi.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1.
The list of dermatophytes used in the study (DOCX 18 kb)
Online Resource 2.
Incidence of dermatophytes, other fungi and yeast in acute patients (DOCX 105 kb)
Online Resource 3
. The viability of Microsporum canis during the co-cultivation with Pythium oligandrum in the liquid suspension (experiment from the Fig. 5) (DOCX 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gabrielová, A., Mencl, K., Suchánek, M. et al. The Oomycete Pythium oligandrum Can Suppress and Kill the Causative Agents of Dermatophytoses. Mycopathologia 183, 751–764 (2018). https://doi.org/10.1007/s11046-018-0277-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-018-0277-2