Abstract
Background
Despite a general decline in mean levels across populations, LDL-cholesterol levels remain a major risk factor for acute coronary syndrome (ACS). The APOB, LDL-R, CILP, and SORT-1 genes have been shown to contain variants that have significant effects on plasma cholesterol levels.
Methods and results
We examined polymorphisms within these genes in 1191 controls and 929 patients with ACS. Only rs646776 within SORT-1 was significantly associated with a risk of ACS (P < 0.05, AA vs. + G comparison; OR 1.21; 95% CI 1.01–1.45). With regard to genetic risk score (GRS), the presence of at least 7 alleles associated with elevated cholesterol levels was connected with increased risk (P < 0.01) of ACS (OR 1.26; 95% CI 1.06–1.52). Neither total mortality nor CVD mortality in ACS subjects (follow up—9.84 ± 3.82 years) was associated with the SNPs analysed or cholesterol-associated GRS.
Conclusions
We conclude that, based on only a few potent SNPs known to affect plasma cholesterol, GRS has the potential to predict ACS risk, but not ACS associated mortality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Increased plasma cholesterol (especially within the LDL fraction), together with smoking, obesity, diabetes, and hypertension, is considered a major contributor to atherosclerotic cardiovascular disease (ACVD) and subsequent acute coronary syndrome (ACS).
Genetic background is an important factor when determining the final values of plasma lipids. Rare monogenic causes of high plasma cholesterol levels have been documented [1]. However, in the majority of subjects, a wide list of genes and variants that exert relatively small but measurable effects can also influence, in addition to unhealthy lifestyles, final plasma lipid concentrations.
The potential association between plasma cholesterol concentration and myocardial infarction was first documented many decades ago [2, 3]. On the one hand, most of the studies that have employed univariable Mendelian randomisation analysis highlight a causality between LDL-cholesterol values and cardiovascular disease [4, 5]. On the other hand, detailed and extensive studies that have utilised the multivariable MR-Egger method have categorically failed to confirm such an association [6, 7]. In fact, several studies suggest that examination of APOB plasma levels is sufficient for risk estimation and, furthermore, that the inclusion of cholesterol values does not further improve risk prediction [6, 8].
For our study, we selected 4 SNPs with a proven and highly significant impact on plasma total cholesterol and LDL-cholesterol in the general Czech population [9]. The first, rs693, is located within APOB (apolipoprotein B, major protein component of LDL particles, OMIM acc. No. 107730); the second, rs16996148, is located at the CILP2/PBX4 loci (cartilage intermediate layer protein 2; OMIM acc. No. 612419 and pre-β-cell leukaemia transcription factor 4; OMIM acc. No. 608127); the third, rs6511720, is located within LDL-R (LDL-receptor, a key protein involved in cholesterol catabolism; OMIM acc. No. 606945); and the fourth, rs646776, is located within the SORT-1 gene (involved in hepatic transport of lipoproteins and arterial calcification; OMIM acc. No. 602458).
The respective importance of the above variants in determining plasma lipid values has been established by independent genome-wide association studies (GWAs) [10,11,12,13] and widely confirmed in different populations. Interestingly, these four SNPs represent just a minority of SNPs (4 out of 26, Hubacek, unpublished results), with confirmed strong effect in Czech general population. Given the polygenic background of hypercholesterolaemia, a genetic risk score (GRS) can be established from these four variants. In different ways, GRSs reflect the simultaneous effect of several individual polymorphisms and are, therefore, believed to be better predictors of disease risk [14, 15] than determinations based on individual genetic variants.
Noteworthy, in observation study protocols, we [16] and others (for example [17, 18]) have recently failed to prove, that increased levels of plasma cholesterol are undoubtedly associated with acute coronary syndrome in different population. This could be partially influenced by the fact, that plasma lipid values generally improved due to the last decades [19] as well as by the fact, that plasma cholesterol is slightly going down after acute coronary syndrome attack [20, 21]. Considering the effect of these variants on plasma cholesterol concentrations, we hypothesized that, if plasma cholesterol remains major traditional risk factor of ACS, individual SNPs and the establishment of a simple cholesterol-determining GRS would reflect an increased risk of ACS in Czech males.
Materials and methods
Subjects
This study included 929 male patients with ACS under the age of 65, consecutively enrolled between April 2006 and February 2015 according to a protocol used at the Cardiology Unit of the Institute for Clinical and Experimental Medicine, Prague (described in detail previously) [22, 23]. Data on mortality [24] were obtained from the Institute of Health, Informatics and Statistics (Ministry of Health of the Czech Republic), where all death certificates are analysed. Reached mean follow-up has been 9.84 ± 3.82 years.
For the control group, 1 191 males aged 25–64 years, all of whom participated in the Czech branch of the post-MONICA study [25]), were selected according to the WHO protocol [26], representing a 1% general population sample from 9 different Czech districts.
To screen for traditional cardiovascular risk factors (cholesterol values, smoking, obesity, hypertension, and diabetes), we used examination procedures described in detail previously [22, 23].
All study participants were self-reported Caucasians.
Genetic analyses
DNA has been isolated from whole EDTA blood samples as described by Miller et al. [27]. Variants within the APOB, CILP2/PBX4, LDL-R and SORT-1 loci were genotyped by PCR–RFLP as described in details before [28]. Briefly, DNA fragment obtaining APOB rs693 variant has been amplified using the 5ʹ aga gga aac caa ggc cac agt tgc and 5ʹ tac att cgg tct cgt gta tct tct oligonucleotides and restriction enzyme XhoI was used to distinguish allele C (uncut PCR product) and T (restriction fragments 110 bp + 26 bp). Oligonucleotides 5ʹ atc cag cta ttt ggg agc agt gtc ctg g and 5ʹ aag gtc tgg tct ctg gaa aac aga ag amplified gene part of SORT-1; restriction enzyme Hin1II produced fragments of 107 bp + 32 bp (allele G) with uncut product of 139 bp representing the allele A. LDL-R has been genotyped by oligonucleotides 5ʹ acc ggg gat gat gat gat tgc and 5ʹ ttg cct aag act tca tta aca ttt g. Alleles G (PCR product of 132 bp) and T (fragments of 106 bp + 26 bp) were distinguished after treatment with enzyme DpnI. The last polymorphism (CILP2/PBX4 loci) has been genotyped with oligonucleotides 5ʹ tgg ctc ttg tcc act ggc cac atc ccc and 5ʹ ttc tcc cat gcc tcc agg ccc cca ag. Restriction enzyme Hin1II produced fragments of 82 bp + 54 bp (allele T) and uncut product of 137 bp is characteristic for the A allele.
Statistical analysis
Chi-square tests and odds ratios (95% CI) were calculated using the freely available Social Science Statistics statistical software package (https://www.socscistatistics.com; accessed February 2022); all procedures used are fully compatible with SPSS software. Cases with fewer than 5 subjects in one category were pooled and analysed together with heterozygotes. A P-value below 0.05 was considered significant.
For calculation of the genetic risk score (GRS), only subjects possessing all 4 SNPs of interest were included (N = 1095 for controls and N = 886 for patients). An unweighted gene score was created for each individual, where subjects received one point for each cholesterol-increasing allele based on associations with plasma cholesterol levels in the general population sample. Due to the low numbers in groups on the opposite end of the score distribution curves, pooling was performed to create GRS sub-groups of “4 or fewer risk alleles” and “7 or more risk alleles”.
As examined SNPs have not been associated with any of the traditional risk factors (prevalence of hypertension, diabetes, smoking status or BMI values) no adjustments were performed.
Results
General characteristics of subjects
The general characteristics of ACS patients and controls are summarized in Table 1. As expected, there were more smokers, diabetics, and hypertensive subjects in the ACS group.
Effects of individual SNPs
In three out of four of the examined SNPs (within the APOB, LDL-R, and CILP/PBX4 loci), overall there was no association between ACS and individual genotypes associated with increased plasma cholesterol values. However, we did detect a slight difference in the case of SORT-1 SNP. AA homozygotes, which have been strongly associated with high plasma levels of total and LDL-cholesterol (P < 0.005) and decreased levels of HDL-cholesterol (P < 0.005) (for more details, see [9]), were slightly more frequent in ACS subjects (64.5% vs. 60.0%; P = 0.03 for AA vs. + G comparison; OR 1.21; 95% CI 1.01–1.45) (Table 2).
Genetic risk score and risk of ACS
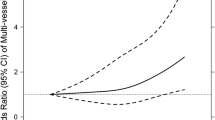
In contrast, the results in respect of GRS were highly significant. As described previously in control subjects [9], total cholesterol and LDL cholesterol increase from those with the lowest to the highest score (P < 0.001 for linear trend). Carriers of 4 or fewer cholesterol-increasing alleles have a significantly lower (P < 0.005) risk of suffering from ACS (OR 0.55; 95% CI 0.37–0.82). Carriers of at least 7 risk alleles are at greater (P < 0.01) risk of ACS (OR 1.26; 95% CI 1.06–1.52) than others (Fig. 1).
Long-term post-ACS mortality
The frequencies of individual SNPs were almost identical (all P-values above 0.44) between ACS survivors and non-survivors (Table 3). GRS was not associated with mortality in CVD subjects (not shown in detail).
Discussion
In our study, Mendelian randomisation (MR) analysis revealed a slight association between plasma cholesterol and increased risk of ACS. With regard to MR, genetic polymorphisms associated with selected biomarkers were used as proxies. Here, four genetic variants were strongly associated with the most frequently cited risk factor of cardiovascular disease (plasma LDL-C levels) and ACS prevalence was dependent (in a slight extent) on genetic risk score established from the four LDL-C associated SNPs.
The importance of individual SNPs in determining anthropometric and/or biochemical parameters is long acknowledged [29,30,31]. However, the vast majority of parameters are determined polygenously and the effect size of individual SNPs is relatively low (albeit functionally significant) and thus represents only minor part in determination of variability of parameters.
For a more nuanced interpretation of the simultaneous effect of several independent genetic variants, different types of genetic risk scores can be created. The respective influences of individual variants at different loci are generally converted into a single value in two ways [14, 15]: an unweighted genetic risk score is the simple sum of risk alleles present in each subject, whereas a weighted genetic risk score is calculated based on the effect size or OR/HR value of each participating SNP, meaning the final value can be time-dependent. Accordingly, absolute values will differ for identical subjects based on age at the time of examination and lifestyle at the time of blood collection.
The importance of GRS is underlined by recent opinions stating that GRS reflects lifetime exposure to risk factors (in the case of our study, to LDL-C levels) and that cumulative load is more precise in risk prediction [32].
Although original GWAs focused on the genetic determination of plasma lipids [10,11,12,13] typically include many thousands of subjects, these results are only useful for clinical purposes when replicated in each particular population. In fact, the effects of about one quarter of variants associated with LDL-cholesterol are not shared between subjects of different ancestry [33], while for triglycerides this “between population” transferability is even lower.
Thus, it is preferable to optimize GRSs for different populations, as not all variants exhibit the same effect in every population [34, 35]. GRS tailoring is especially necessary in cases where it is employed to predict diseases of affluence such as cardiovascular conditions, diabetes, and obesity. In these instances, a substantial proportion of risk is attributable to unhealthy lifestyle factors. The resulting potential inter-population differences can also be associated with gene-environment interactions. In such cases, the identical genetic variant can exert a variety of effects in different [14], or even in identical [36], populations according to environmental conditions.
Our previous analysis [9] clearly confirmed that all four GWAs-retrieved SNPs had a significant effect on plasma LDL-C levels in the Czech population. The effect size was between approximately 5% and 13% for individual SNPs. Both LDL-C and TC were higher by ~ 0.5 mmol/L for carriers of 8 risk alleles than for carriers of 4 or fewer risk alleles, a finding comparable with several previous studies. For example, one report found that a GRS established based on 23 SNPs was associated with TC levels between ~ 5.2 mmol/L (carriers of 11 or fewer cholesterol-raising alleles) and ~ 6.0 mmol/L for subjects with 18 or more alleles [37]. Shahid et al. found atherogenic blood lipids to have a significant positive association with GRS [38]. Their score, derived from 21 SNPs, revealed TC values ranging from ~ 4.7 mmol/L in carriers of 14 or fewer cholesterol-raising alleles to ~ 5.7 mmol/L in carriers with 21 or more risk alleles. An analysis of two UK biobank studies (WHII and BWHHS) [39] demonstrated differences between opposite GRS quintiles ranging from 0.6 to 0.9 mmol/L for both LDL-C and TC. Interestingly, the authors used different (albeit significantly overlapping) sets of SNPs to determine TC (20 SNPs) and LDL-C (22 SNPs). Finally, almost 8 500 SNPs were used to establish a GRS for White British UK Biobank participants, accounting for over 20% of the variance in LDL-C concentration [40].
As recently underlined GRS are important [41], but not all-explanatory [42] tools to predict medical events. Importantly—we believe, that the analysis of genetic predisposition to any noncommunicable disease needs to be (to be clinically useful) performed in time (at young age, probably at 25 years latest) and before the any onset of traditional risk factors. This was the secondary reason not to adjust our results for potential confounding factors. At this age, genetic predisposition could point on subjects under risk. Using the tools as intensive lifestyle intervention and more intensive and more focused screening programs, diseases onset could be, if not fully avoided, at least postponed to higher age categories.
Similar to screening for FH-causing mutations, screening for several common variants combined with a simultaneous/cumulative analysis of their effects can clearly identify potential patients at increased risk of hypercholesterolaemia and subsequent cardiovascular disease [43]. Nonetheless, it should be noted that the abovementioned studies significantly differed not only in terms of the number of SNPs but also in the method of selection.
There are uncertainties concerning the ideal number of variants to include in GRS calculations. A wide list of variants with only subtle effect sizes can in fact compromise the accuracy of results. The numbers of examined subjects used to identify risk allele/effect size are time-dependent and affected by potential selection bias. The smaller the cholesterol-raising effect size detected, the greater the chance of false-positive results. The actual effect across an entire population may be in fact zero or even negative. The “over-inclusion” of these variants in GRS calculations also results in a low cost–benefit. For example, Khera et al. [44] analysed an extreme 6.6 million SNPs to quantify CVD risk, where the top 5% of subjects with the highest GRS had “only” a 3.7-fold increased risk (nominally 17.5% of patients fell within this category compared to 5% within the rest of the population) of early onset myocardial infarction in comparison with other individuals. Shahid et al. [38] documented only a slightly lower OR (2.96) using a GRS based on a mere 21 SNPs.
Despite focusing on a different lifestyle-associated disease, namely type 2 diabetes mellitus, one study, which used results from the Estonian Biobank cohort, demonstrated that a GRS established from a maximum of 1000 SNPs is a better predictor of disease than a score established from a higher number of SNPs [45].
Two secondary outcomes of our study are noteworthy. Firstly, genotypes associated in the general population with increased plasma LDL-cholesterol values were more frequent than genotypes associated with lower plasma LDL-cholesterol values. Thus, it can be assumed that selection pressure during the ancient era of human development resulted in the “promotion” of these alleles. It is probable that one of the benefits of the increase in plasma cholesterol levels over time was to counteract infection [46]. The authors of a Dutch study have documented the advantage of higher total cholesterol in a historical context, finding that carriers of FH-causing mutations lived significantly longer until about the end of the nineteenth century than non-carriers [47].
Secondly, genotypes associated with increased plasma LDL-C values in the general population were only very slightly (and mostly non-significantly) over-represented in patients with ACVD compared to controls. Although we found a continuous increase in plasma LDL-C values in subjects along with a sequentially increasing number of risk alleles, only the subgroup comprising individuals with at least 7 risk/cholesterol-increasing alleles, i.e. in subjects with really high plasma cholesterol levels, were at increased risk of ACS. Nonetheless, although there was a trend toward higher GRS values in patients, the differences between patients and controls were more evident at the opposite end of the distribution curve, characterized by low GRS values.
In conclusion, our study indicates that genetic risk score, based on only a few individual SNPs, is a significant predictor of acute coronary syndrome in the Czech population even in cases where individual SNPs are associated with plasma cholesterol but not with increased risk of ACS per se. Importantly predisposition to low plasma cholesterol levels seems to be of greater importance than a predisposition to increased levels.
Data availability
Raw data are available for collaboration purposes upon request at corresponding author.
Code availability
Not applicable.
References
Vrablik M, Tichý L, Freiberger T, Blaha V, Satny M, Hubacek JA (2020) Genetics of familial hypercholesterolemia: new insights. Front Genet 11:574474. https://doi.org/10.3389/fgene.2020.574474
LaLiberte H, Vachon M (1948) Infarctus du myocarde et cholestérol; rapport préliminaire (Myocardial infarction and cholesterol; preliminary report). Laval Med 13(3):294–302
Lm M, Hall L, Al C (1948) Cholesterol and cholesterol ester levels in acute myocardial infarction. Am Heart J 35(5):866. https://doi.org/10.1016/0002-8703(48)90536-5
Holmes MV, Asselbergs FW, Palmer TM, Drenos F, Lanktree MB, Nelson CP et al (2015) Mendelian randomization of blood lipids for coronary heart disease. Eur Heart J 36(9):539–550. https://doi.org/10.1093/eurheartj/eht571
Allara E, Morani G, Carter P, Gkatzionis A, Zuber V, Foley CN et al (2019) Genetic determinants of lipids and cardiovascular disease outcomes: a wide-angled Mendelian randomization investigation. Circ Genom Precis Med 12(12):e002711. https://doi.org/10.1161/CIRCGEN.119.002711
Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, Holmes MV (2020) Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med 17(3):e1003062. https://doi.org/10.1371/journal.pmed.1003062
He L, Culminskaya I, Loika Y, Arbeev KG, Bagley O, Duan M, Yashin AI, Kulminski AM (2018) Causal effects of cardiovascular risk factors on onset of major age-related diseases: a time-to-event Mendelian randomization study. Exp Gerontol 107:74–86. https://doi.org/10.1016/j.exger.2017.09.019
Ference BA, Kastelein JJP, Ray KK, Ginsberg HN, Chapman MJ, Packard CJ et al (2019) Association of triglyceride-lowering LPL variants and LDL-C-lowering LDLR variants with risk of coronary heart disease. JAMA 321(4):364–373. https://doi.org/10.1001/jama.2018.20045
Hubacek JA, Adamkova V, Lanska V, Dlouha D (2017) Polygenic hypercholesterolemia: examples of GWAS results and their replication in the Czech-Slavonic population. Physiol Res 66(Suppl 1):S101–S111. https://doi.org/10.33549/physiolres.933580
Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE et al (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41(1):56–65. https://doi.org/10.1038/ng.291
Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP et al (2009) Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 41(1):47–55. https://doi.org/10.1038/ng.269
Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M et al (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466(7307):707–713. https://doi.org/10.1038/nature09270
Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S et al (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45(11):1274–1283. https://doi.org/10.1038/ng.2797
Osterman MD, Kinzy TG, Cooke Bailey JN (2021) Polygenic risk scores. Curr Protoc 1(5):e126. https://doi.org/10.1002/cpz1.126. Erratum in: (2022) Curr Protoc 2:e553
Lewis CM, Vassos E (2020) Polygenic risk scores: from research tools to clinical instruments. Genome Med 12:44. https://doi.org/10.1186/s13073-020-00742-5
Hubacek JA, Stanek V, Gebauerova M, Adamkova V, Lesauskaite V, Zaliaduonyte-Peksiene D et al (2017) Traditional risk factors of acute coronary syndrome in four different male populations—total cholesterol value does not seem to be relevant risk factor. Physiol Res 66(Suppl 1):S121–S128. https://doi.org/10.33549/physiolres.933597
Guan D, Ji Y, Lu X, Feng W, Ma W (2023) Associations of MTHFR gene polymorphism with lipid metabolism and risk of cerebral infarction in the Northwest Han Chinese population. Front Neurol 14:1152351. https://doi.org/10.3389/fneur.2023.1152351
Škrlec I, Biloglav Z, Talapko J, Džijan S, Daus-Šebeđak D, Cesar V (2023) Myocardial infarction susceptibility and the MTNR1B polymorphisms. Int J Mol Sci 24:11444. https://doi.org/10.3390/ijms241411444
Cífková R, Bruthans J, Wohlfahrt P, Hrubeš Krajčoviechová A, Šulc P, Jozífová M et al (2023) Longitudinal trends in severe dyslipidemia in the Czech population: the Czech MONICA and Czech post-MONICA study. J Cardiovasc Dev Dis 10:328. https://doi.org/10.3390/jcdd10080328
Fresco C, Maggioni AP, Signorini S, Merlini PA, Mocarelli P, Fabbri G et al (2002) Variations in lipoprotein levels after myocardial infarction and unstable angina: the LATIN trial. Ital Heart J 3:587–592
Wattanasuwan N, Khan IA, Gowda RM, Vasavada BC, Sacchi TJ (2001) Effect of acute myocardial infarction on cholesterol ratios. Chest 120:1196–1199. https://doi.org/10.1378/chest.120.4.1196
Pitha J, Hubacek JA, Poledne R, Staněk V, Aschermann M, Gebauerová M et al (2007) Genetic determination of the prognosis in survivors of acute coronary syndromes. Study design and rationale for a multicenter study. Cor Vasa 49:134–137
Hubacek JA, Vrablik M, Dlouha D, Stanek V, Gebauerova M, Adamkova V et al (2016) Gene variants at FTO, 9p21, and 2q36.3 are age-independently associated with myocardial infarction in Czech men. Clin Chim Acta 454:119–123. https://doi.org/10.1016/j.cca.2016.01.005
Hubacek JA, Staněk V, Gebauerová M, Poledne R, Aschermann M, Skalická H et al (2015) Rs6922269 marker at the MTHFD1L gene predict cardiovascular mortality in males after acute coronary syndrome. Mol Biol Rep 42(8):1289–1293. https://doi.org/10.1007/s11033-015-3870-1
Cífková R, Skodová Z, Bruthans J, Adámková V, Jozífová M, Galovcová M et al (2010) Longitudinal trends in major cardiovascular risk factors in the Czech population between 1985 and 2007/8. Czech MONICA and Czech post-MONICA. Atherosclerosis 211(2):676–681. https://doi.org/10.1016/j.atherosclerosis.2010.04.007
Tunstall-Pedoe H, Kuulasmaa K, Tolonen H et al (2003) MONICA Monograph and multimedia sourcebook: world‘s largest study of heart disease, stroke, risk factors, and population trends 1979–2002. World Health Organization, Geneva
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for DNA extraction from human nucleated cells. Nucleic Acid Res 16:1215. https://doi.org/10.1093/nar/16.3.1215
Vrablík M, Hubáček JA, Dlouhá D, Lánská V, Rynekrová J, Zlatohlávek L, Prusíková M, Ceška R, Adámková V (2012) Impact of variants within seven candidate genes on statin treatment efficacy. Physiol Res 61(6):609–617. https://doi.org/10.33549/physiolres.932341
Hubacek JA (2016) Apolipoprotein A5 fifteen years anniversary: lessons from genetic epidemiology. Gene 592(1):193–199. https://doi.org/10.1016/j.gene.2016.07.070
Yılmaz B, Gezmen KM (2020) The current review of adolescent obesity: the role of genetic factors. J Pediatr Endocrinol Metab 34(2):151–162. https://doi.org/10.1515/jpem-2020-0480
SEARCH Collaborative Group, Link E, Parish S, Armitage J, Bowman L, Heath S et al (2008) SLCO1B1 variants and statin-induced myopathy—a genomewide study. N Engl J Med 359(8):789–799. https://doi.org/10.1056/NEJMoa0801936
Shapiro MD, Bhatt DL (2020) “Cholesterol-years” for ASCVD risk prediction and treatment. J Am Coll Cardiol 76:1517–1520. https://doi.org/10.1016/j.jacc.2020.08.004
Kuchenbaecker K, Telkar N, Reiker T, Walters RG, Lin K, Eriksson A et al (2019) The transferability of lipid loci across African, Asian and European cohorts. Nat Commun 10(1):4330. https://doi.org/10.1038/s41467-019-12026-7
Janssens ACJW (2019) Validity of polygenic risk scores: are we measuring what we think we are? Hum Mol Genet 28(R2):R143–R150. https://doi.org/10.1093/hmg/ddz205
Hubacek JA, Dlouha D, Adamkova V, Schwarzova L, Lanska V, Ceska R et al (2019) The gene score for predicting hypertriglyceridemia: new insights from a Czech case-control study. Mol Diagn Ther 23:555–562. https://doi.org/10.1007/s40291-019-00412-2
Hubacek JA, Pitha J, Skodová Z, Poledne R, Lánská V, Waterworth DM et al (2003) Polymorphisms in CYP-7A1, not APOE, influence the change in plasma lipids in response to population dietary change in an 8 year follow-up; results from the Czech MONICA study. Clin Biochem 36:263–267. https://doi.org/10.1016/s0009-9120(03)00025-0
Lu Y, Feskens EJ, Boer JM, Imholz S, Verschuren WM, Wijmenga C et al (2010) Exploring genetic determinants of plasma total cholesterol levels and their predictive value in a longitudinal study. Atherosclerosis 213(1):200–205. https://doi.org/10.1016/j.atherosclerosis.2010.08.053
Shahid SU, Shabana CJA, Beaney KE, Li K, Rehman A, Humphries SE (2017) Genetic risk analysis of coronary artery disease in Pakistani subjects using a genetic risk score of 21 variants. Atherosclerosis 258:1–7. https://doi.org/10.1016/j.atherosclerosis.2017.01.024
Shah S, Casas JP, Gaunt TR, Cooper J, Drenos F, Zabaneh D et al (2013) Influence of common genetic variation on blood lipid levels, cardiovascular risk, and coronary events in two British prospective cohort studies. Eur Heart J 34(13):972–981. https://doi.org/10.1093/eurheartj/ehs243
Wu H, Forgetta V, Zhou S, Bhatnagar SR, Paré G, Richards JB (2021) Polygenic risk score for low-density lipoprotein cholesterol is associated with risk of ischemic heart disease and enriches for individuals with familial hypercholesterolemia. Circ Genom Precis Med 14(1):e003106. https://doi.org/10.1161/CIRCGEN.120.003106
Kavousi M, Schunkert H (2022) Polygenic risk score: a tool ready for clinical use? Eur Heart J 43:1712–1714. https://doi.org/10.1093/eurheartj/ehab923
Hingorani AD, Gratton J, Finan C, Schmidt AF, Patel R, Sofat R, Kuan V et al (2023) Performance of polygenic risk scores in screening, prediction, and risk stratification: secondary analysis of data in the polygenic score catalog. BMJ Med 2:e000554. https://doi.org/10.1136/bmjmed-2023-000554
Futema M, Taylor-Beadling A, Williams M, Humphries SE (2021) Genetic testing for familial hypercholesterolemia-past, present, and future. J Lipid Res 62:100139. https://doi.org/10.1016/j.jlr.2021.100139
Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P et al (2019) Whole-genome sequencing to characterize monogenic and polygenic contributions in patients hospitalized with early-onset myocardial infarction. Circulation 139(13):1593–1602. https://doi.org/10.1161/CIRCULATIONAHA.118.035658
Läll K, Mägi R, Morris A, Metspalu A, Fischer K (2017) Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet Med 19(3):322–329. https://doi.org/10.1038/gim.2016.103
Poledne R, Zicha J (2018) Human genome evolution and development of cardiovascular risk factors through natural selection. Physiol Res 67(2):155–163. https://doi.org/10.33549/physiolres.933885
Sijbrands EJ, Westendorp RG, Defesche JC, de Meier PH, Smelt AH, Kastelein JJ (2001) Mortality over two centuries in large pedigree with familial hypercholesterolaemia: family tree mortality study. BMJ 322(7293):1019–1023. https://doi.org/10.1136/bmj.322.7293.1019
Funding
Open access publishing supported by the National Technical Library in Prague. This study was supported by Ministry of Health, Czech Republic—Conceptual Development of Research Organisation (Institute for Clinical and Experimental Medicine—IKEM, IN 00023001) and by the project National Institute for Research of Metabolic and Cardiovascular Diseases (Program EXCELES, Project No. LX22NPO5104)—Funded by the European Union—Next Generation EU.
Author information
Authors and Affiliations
Contributions
Conceptualisation, JAH, VA, VS, JP, JKa, JKe; methodology, JAH, VA, VS, JKa, JKe, JP; formal analysis, VL, JAH; investigation, JP, VS, VA, JM; resources, JP, VS, VA, JM; VL, JM, JKa, JKe, JAH; writing—original draft preparation, JAH, JP; writing—review and editing, VA, VL, VS, JM, MG, JKa, JKe, JP; supervision, JP, JM, MG, VS; project administration, VL, JM; funding acquisition, JAH, JP All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Institutional Review Board of the Institute for Clinical and Experimental medicine in accordance with the 1964 Declaration of Helsinki.
Informed consent
All subjects signed informed consent with the participation in the study.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hubacek, J.A., Adamkova, V., Lanska, V. et al. Cholesterol associated genetic risk score and acute coronary syndrome in Czech males. Mol Biol Rep 51, 164 (2024). https://doi.org/10.1007/s11033-023-09128-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-09128-3