Abstract
Background
Multiple sclerosis (MS) is a progressive neurodegenerative disease of the central nervous system (CNS) with varying degrees of axonal and neuronal damage. The onset and progression of the disease are influenced by several environmental and genetic variables. Long non-coding RNAs (lncRNAs) have a crucial role in the pathophysiology of MS. Our study aimed to assess the levels of HAR1A and HAR1B lncRNA expression in the blood samples of MS patients and investigate the relationship between these lncRNAs and disease activity.
Methods and results
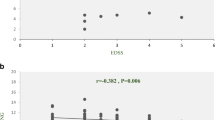
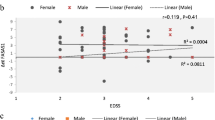
The blood samples of 100 MS patients, including 82 relapsing-remitting (RR), 8 primary progressive (PP), and 10 secondary progressive (SP) MS cases, and 100 healthy controls were collected. Quantitative real-time PCR was used for the evaluation of gene expression. ROC curve analysis was performed to evaluate the diagnostic potential of lncRNA levels. A significant decrease was detected in HAR1A expressions (P < 0.0001), and a moderate increase was also shown in HAR1B of SPMS patients (P value = 0.0189). HAR1A showed different expression levels in patients over forty (P value = 0.034). The expression levels of HAR1A and HAR1B were positively correlated in MS patients (r = 0.2003, P value = 0.0457). In addition, ROC curve results suggested that HAR1A can be introduced as a novel biomarker for MS diagnosis (AUC = 0.776).
Conclusion
The low serum level of HAR1A may be a potential molecular biomarker for MS diagnosis; however, no discernible difference was detected in the expression level of HAR1B in the blood samples of MS patients.




Similar content being viewed by others
Data availability
Data are available upon request.
Code availability
Not applicable.
References
Sedighi S, Gholizadeh O, Yasamineh S, Akbarzadeh S, Amini P, Favakehi P, Afkhami H, Firouzi-Amandi A, Pahlevan D, Eslami M, Yousefi B, Poortahmasebi V, Dadashpour M (2022) Comprehensive Investigations relationship between viral infections and multiple sclerosis pathogenesis. Curr Microbiol 80(1):15. https://doi.org/10.1007/s00284-022-03112-z
Walton C, King R, Rechtman L, Kaye W, Leray E, Marrie RA, Robertson N, La Rocca N, Uitdehaag B, van der Mei I (2020) Rising prevalence of multiple sclerosis worldwide: insights from the Atlas of MS. Multiple Scler J 26(14):1816–1821
Asmarian N, Sharafi Z, Mousavi A, Jacques R, Tamayo I, Bind M-A, Abutorabi-Zarchi M, Moradian MJ, Izadi S (2021) Multiple sclerosis incidence rate in southern Iran: a bayesian epidemiological study. BMC Neurol 21(1):1–9
Choi I-Y, Lee P, Adany P, Hughes AJ, Belliston S, Denney DR, Lynch SG (2018) In vivo evidence of oxidative stress in brains of patients with progressive multiple sclerosis. Multiple Scler J 24(8):1029–1038
Fenoglio C, Oldoni E, Serpente M, Milena A, Arcaro M, D’Anca M, Pietroboni AM, Calvi A, Lecchi E, Goris A (2018) LncRNAs expression profile in peripheral blood mononuclear cells from multiple sclerosis patients. J Neuroimmunol 324:129–135
Amatya B, Khan F, Galea M (2019) Rehabilitation for people with multiple sclerosis: an overview of Cochrane Reviews. Cochrane Database Syst Rev. https://doi.org/10.1002/14651858.CD012732.pub2
Fortune AJ, Taylor BV, Charlesworth JC, Burdon KP, Blackburn NB, Fletcher JL, Mehta A, Young KM (2022) Generation and characterisation of four multiple sclerosis iPSC lines from a single family. Stem Cell Res 62:102828
Krysko KM, Graves JS, Dobson R, Altintas A, Amato MP, Bernard J, Bonavita S, Bove R, Cavalla P, Clerico M (2020) Sex effects across the lifespan in women with multiple sclerosis. Ther Adv Neurol Disord 13:1756286420936166
Bar-Or A, Pender MP, Khanna R, Steinman L, Hartung H-P, Maniar T, Croze E, Aftab BT, Giovannoni G, Joshi MA (2020) Epstein–Barr virus in multiple sclerosis: theory and emerging immunotherapies. Trends Mol Med 26(3):296–310
Cencioni MT (2020) The immune regulation of PD-1/PDL-1 axis, a potential biomarker in multiple sclerosis. Neuroimmunol Neuroinflamm 7(3):277–290
Chan VS-F (2020) Epigenetics in multiple sclerosis. In: Epigenetics in allergy and autoimmunity, pp 309–374
Zhang F, Gao C, Ma XF, Peng XL, Zhang RX, Kong DX, Simard AR, Hao JW (2016) Expression Profile of Long Noncoding RNA s in Peripheral Blood mononuclear cells from multiple sclerosis patients. CNS Neurosci Ther 22(4):298–305
Santoro M, Nociti V, Lucchini M, De Fino C, Losavio FA, Mirabella M (2016) Expression profile of long non-coding RNAs in serum of patients with multiple sclerosis. J Mol Neurosci 59(1):18–23
Statello L, Guo C-J, Chen L-L, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22(2):96–118
Ozdilek B, Kaya IA, Demircan B, Tombul T, Ankarali H (2021) The relationship between serum levels of lncRNA H19, GAS5, HAR1B, LINC01783 and clinical signs of Parkinson’s disease
Miao C, Bai L, Yang Y, Huang J (2021) Dysregulation of lncRNAs in rheumatoid arthritis: biomarkers, pathogenesis and potential therapeutic targets. Front Pharmacol 12:652751
Gao Y, Li S, Zhang Z, Yu X, Zheng J (2018) The role of long non-coding RNAs in the pathogenesis of RA, SLE, and SS. Front Med 5:193
Doxtater K, Tripathi MK, Khan MM (2020) Recent advances on the role of long non-coding RNAs in Alzheimer’s disease. Neural Regen Res 15(12):2253
Eftekharian MM, Ghafouri-Fard S, Soudyab M, Omrani MD, Rahimi M, Sayad A, Komaki A, Mazdeh M, Taheri M (2017) Expression analysis of long non-coding RNAs in the blood of multiple sclerosis patients. J Mol Neurosci 63(3):333–341
Amiri M, Mokhtari MJ, Bayat M, Safari A, Dianatpuor M, Tabrizi R, Borhani-Haghighi A (2022) Expression and diagnostic values of MIAT, H19, and NRON long non-coding RNAs in multiple sclerosis patients. Egypt J Med Hum Genet 23(1):1–9
Shaker OG, Mahmoud RH, Abdelaleem OO, Ibrahem EG, Mohamed AA, Zaki OM, Abdelghaffar NK, Ahmed TI, Hemeda NF, Ahmed NA (2019) LncRNAs, MALAT1 and lnc-DC as potential biomarkers for multiple sclerosis diagnosis. Biosci Rep 39 (1)
Nociti V, Santoro M (2021) What do we know about the role of lncRNAs in multiple sclerosis? Neural Regen Res 16(9):1715
Dastmalchi R, Ghafouri-Fard S, Omrani MD, Mazdeh M, Sayad A, Taheri M (2018) Dysregulation of long non-coding RNA profile in peripheral blood of multiple sclerosis patients. Mult Scler Relat Disord 25:219–226
Mourtada-Maarabouni M, Hasan AM, Farzaneh F, Williams GT (2010) Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Mol Pharmacol 78(1):19–28
Ziegeler M, Cevec M, Richter C, Schwalbe H (2012) NMR studies of HAR1 RNA secondary structures reveal conformational dynamics in the human RNA. ChemBioChem 13(14):2100–2112
Pollard KS, Salama SR, Lambert N, Lambot M-A, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A (2006) An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443(7108):167–172
Waters E, Pucci P, Hirst M, Chapman S, Wang Y, Crea F, Heath CJ (2021) HAR1: an insight into lncRNA genetic evolution. Epigenomics 13(22):1831–1843
Salviano-Silva A, Lobo-Alves SC, Almeida RCd, Malheiros D, Petzl-Erler ML (2018) Besides pathology: long non-coding RNA in cell and tissue homeostasis. Non-coding RNA 4(1):3
Talebian S, Gharesouran J, Ghafouri-Fard S, Esfahani BS, Arsang-Jang S, Omrani MD, Taheri M, Rezazadeh M (2019) Assessment of expression of RELN signaling pathway in multiple sclerosis patients. Immunobiology 224(3):402–407
Wang K, Song F, Fernandez-Escobar A, Luo G, Wang J-H, Sun Y (2018) The properties of cytokines in multiple sclerosis: pros and cons. Am J Med Sci 356(6):552–560
Lee C-P, Ko AM-S, Nithiyanantham S, Lai C-H, Ko Y-C (2021) Long noncoding RNA HAR1A regulates oral cancer progression through the alpha-kinase 1, bromodomain 7, and myosin IIA axis. J Mol Med 99(9):1323–1334
Jafari A, Babajani A, Rezaei-Tavirani M (2021) Multiple sclerosis biomarker discoveries by proteomics and metabolomics approaches. Biomark Insights 16:11772719211013352
Johnson R, Richter N, Jauch R, Gaughwin PM, Zuccato C, Cattaneo E, Stanton LW (2010) Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol Genom 41(3):269–274
Nopoulos PC (2022) Huntington disease: a single-gene degenerative disorder of the striatum. Dialogues Clin Neurosci 19:91
Sunwoo J-S, Lee S-T, Im W, Lee M, Byun J-I, Jung K-H, Park K-I, Jung K-Y, Lee SK, Chu K (2017) Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol Neurobiol 54(2):1577–1586
Tolosa A, Sanjuan J, Leal C, Costas J, Molto M, De Frutos R (2008) Rapid evolving RNA gene HAR1A and schizophrenia. Schizophr Res 99(1–3):370–372
Chen X, Yan G-Y (2013) Novel human lncRNA–disease association inference based on lncRNA expression profiles. Bioinformatics 29(20):2617–2624
Chen X, Yan CC, Zhang X, You Z-H (2017) Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform 18(4):558–576
Costaa MBW, zu Siederdissenc CH, Tulpand D, Stadlerc P, Nowicka K. Uncovering the structural evolution of the human accelerated region
Armstrong NC, Anderson RC, McDermott KW (2019) Reelin: diverse roles in central nervous system development, health and disease. Int J Biochem Cell Biol 112:72–75
Chen Y, Guo Y, Chen H, Ma F (2020) Long non-coding RNA expression profiling identifies a four-long non-coding RNA prognostic signature for isocitrate dehydrogenase mutant glioma. Front Neurol 11:573264
Verdelli C, Morotti A, Tavanti GS, Silipigni R, Guerneri S, Ferrero S, Vicentini L, Vaira V, Corbetta S (2021) The core stem genes SOX2, POU5F1/OCT4, and NANOG are expressed in human parathyroid tumors and modulated by MEN1, YAP1, and β-catenin pathways activation. Biomedicines 9(6):637
Dolati S, Ahmadi M, Aghebti-Maleki L, Nikmaram A, Marofi F, Rikhtegar R, Ayromlou H, Yousefi M (2018) Nanocurcumin is a potential novel therapy for multiple sclerosis by influencing inflammatory mediators. Pharmacol Rep 70(6):1158–1167
Acknowledgements
The authors would like to thank the Department of Animal Biology, University of Tabriz, for supporting this project.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author information
Authors and Affiliations
Contributions
SA: Conceptualization, Visualization, Writing—original draft, Writing—review and editing, investigation. ST-G: Conceptualization, Visualization, investigation. PN: investigation. AR: Conceptualization, review and editing, Formal analysis. TG: Writing—review and editing. MAHF: Review and editing, Supervision. RS: Visualization, Supervision
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
The Medical Ethics Committee of Tabriz Medical University approved the study (Approval Number: ID No.IR.TABRIZU.REC.1401.034).
Consent to participate
Written informed consent was obtained from all patients.
Consent for publication
All authors have given their consent for publication in this journal.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Akbarzadeh, S., Tayefeh-Gholami, S., Najari, P. et al. The expression profile of HAR1A and HAR1B in the peripheral blood cells of multiple sclerosis patients. Mol Biol Rep 50, 2391–2398 (2023). https://doi.org/10.1007/s11033-022-08182-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08182-7