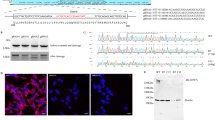
Abstract
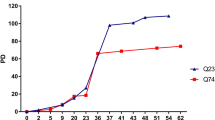
Huntington’s disease (HD) is a devastating neurodegenerative disease caused by cytosine-adenine-guanine trinucleotide repeat expansion in the huntingtin gene. Growing evidence supports the regulatory functions of long noncoding RNAs (lncRNAs) in the disease process, but little is known about the association between lncRNAs and neuronal death in HD. Here, we evaluated the altered expression profiles of lncRNA in HD by using microarrays. Among dysregulated lncRNAs, we focused on the upregulation of nuclear paraspeckle assembly transcript 1 (NEAT1). Quantitative PCR analysis validated increased NEAT1 levels in the R6/2 mouse brain as well as the human HD postmortem brain. To determine the biological effects of NEAT1 on neuronal survival, neuro2A cells were transfected with the NEAT1 short isoform vector and were subjected to H2O2-induced injury. Subsequently, NEAT1-transfected cells showed increased viability under oxidative stress. Our observations support the notion that NEAT1 upregulation in HD contributes to the neuroprotective mechanism against neuronal injury rather than the pathological process underlying neurodegeneration in HD.


Similar content being viewed by others
References
Labbadia J, Morimoto RI (2013) Huntington’s disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci 38:378–385. doi:10.1016/j.tibs.2013.05.003
MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA et al (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983. doi:10.1016/0092-8674(93)90585-E
Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J et al (2006) Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell 125:1179–1191. doi:10.1016/j.cell.2006.04.026
Sugars KL, Rubinsztein DC (2003) Transcriptional abnormalities in Huntington disease. Trends Genet 19:233–238. doi:10.1016/S0168-9525(03)00074-X
Trushina E, McMurray CT (2007) Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience 145:1233–1248. doi:10.1016/j.neuroscience.2006.10.056
Lee ST, Chu K, Jung KH, Im WS, Park JE, Lim HC, Won CH, Shin SH et al (2009) Slowed progression in models of Huntington disease by adipose stem cell transplantation. Ann Neurol 66:671–681. doi:10.1002/ana.21788
Ross CA, Tabrizi SJ (2011) Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol 10:83–98. doi:10.1016/S1474-4422(10)70245-3
Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. doi:10.1146/annurev-biochem-051410-092902
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47:199–208. doi:10.1038/ng.3192
Kung JTY, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193:651–669. doi:10.1534/genetics.112.146704
Wahlestedt C (2013) Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 12:433–446. doi:10.1038/nrd4018
Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd et al (2008) Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14:723–730. doi:10.1038/nm1784
Lee DY, Moon J, Lee ST, Jung KH, Park DK, Yoo JS, Sunwoo JS, Byun JI et al (2015) Distinct expression of long non-coding RNAs in an Alzheimer’s disease model. J Alzheimers Dis 45:837–849. doi:10.3233/jad-142919
Lee DY, Moon J, Lee ST, Jung KH, Park DK, Yoo JS, Sunwoo JS, Byun JI et al (2015) Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem Biophys Res Commun 462:433–440. doi:10.1016/j.bbrc.2015.04.149
Johnson R, Richter N, Jauch R, Gaughwin PM, Zuccato C, Cattaneo E, Stanton LW (2010) Human accelerated region 1 noncoding RNA is repressed by REST in Huntington’s disease. Physiol Genomics 41:269–274. doi:10.1152/physiolgenomics.00019.2010
Chung DW, Rudnicki DD, Yu L, Margolis RL (2011) A natural antisense transcript at the Huntington’s disease repeat locus regulates HTT expression. Hum Mol Genet 20:3467–3477. doi:10.1093/hmg/ddr263
Francelle L, Galvan L, Gaillard MC, Petit F, Bernay B, Guillermier M, Bonvento G, Dufour N, Elalouf JM, Hantraye P, Deglon N, de Chaldee M, Brouillet E (2015) Striatal long noncoding RNA Abhd11os is neuroprotective against an N-terminal fragment of mutant huntingtin in vivo. Neurobiol Aging 36:1601.e1607-1616. doi:10.1016/j.neurobiolaging.2014.11.014
Johnson R (2012) Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis 46:245–254. doi:10.1016/j.nbd.2011.12.006
Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T (2009) MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 106:2525–2530. doi:10.1073/pnas.0807899106
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726. doi:10.1016/j.molcel.2009.01.026
Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL (2009) MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19:347–359. doi:10.1101/gr.087775.108
Fox AH, Lamond AI (2010) Paraspeckles. Cold Spring Harb Perspect Biol 2:a000687. doi:10.1101/cshperspect.a000687
Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39. doi:10.1186/1471-2164-8-39
Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J et al (2014) The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 5:5383. doi:10.1038/ncomms6383
Chen X, Kong J, Ma Z, Gao S, Feng X (2015) Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res 5:2808–2815
He C, Jiang B, Ma J, Li Q (2015) Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. APMIS. doi:10.1111/apm.12480
Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F, Zhuo C, Zheng Y et al (2015) Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer 14:191. doi:10.1186/s12943-015-0455-5
Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y et al (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87:493–506
Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH et al (2011) Altered microRNA regulation in Huntington’s disease models. Exp Neurol 227:172–179. doi:10.1016/j.expneurol.2010.10.012
Zhang Q, Chen CY, Yedavalli VS, Jeang KT (2013) NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 4:e00596–00512. doi:10.1128/mBio.00596-12
Kim M, Lee HS, LaForet G, McIntyre C, Martin EJ, Chang P, Kim TW, Williams M et al (1999) Mutant huntingtin expression in clonal striatal cells: dissociation of inclusion formation and neuronal survival by caspase inhibition. J Neurosci 19:964–973
Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME (2000) Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet 9:2799–2809. doi:10.1093/hmg/9.19.2799
Naganuma T, Hirose T (2013) Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol 10:456–461. doi:10.4161/rna.23547
Chen LL, Carmichael GG (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35:467–478. doi:10.1016/j.molcel.2009.06.027
Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y et al (2014) Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 53:393–406. doi:10.1016/j.molcel.2014.01.009
Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S et al (2014) NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 25:169–183. doi:10.1091/mbc.E13-09-0558
Saha S, Murthy S, Rangarajan PN (2006) Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol 87:1991–1995. doi:10.1099/vir.0.81768-0
Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ (2011) Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem 116:459–466. doi:10.1111/j.1471-4159.2010.07126.x
Nishimoto Y, Nakagawa S, Hirose T, Okano HJ, Takao M, Shibata S, Suyama S, Kuwako K et al (2013) The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain 6:31. doi:10.1186/1756-6606-6-31
Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H et al (2007) Global changes to the ubiquitin system in Huntington’s disease. Nature 448:704–708. doi:10.1038/nature06022
Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Kruger E (2003) Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem 278:21517–21525. doi:10.1074/jbc.M301032200
Shav-Tal Y, Zipori D (2002) PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett 531:109–114. doi:10.1016/S0014-5793(02)03447-6
Dong X, Sweet J, Challis JR, Brown T, Lye SJ (2007) Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol Cell Biol 27:4863–4875. doi:10.1128/MCB.02144-06
Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, Hayward SD, Moran TH et al (2005) p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron 47:29–41. doi:10.1016/j.neuron.2005.06.005
Johnson R, Buckley NJ (2009) Gene dysregulation in Huntington’s disease: REST, microRNAs and beyond. Neuromolecular Med 11:183–199. doi:10.1007/s12017-009-8063-4
Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schodel J, Green CM et al (2015) Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34:4482–4490. doi:10.1038/onc.2014.378
Li Z, Wang L, Wang Y, Liu L, Wang L, Li W, Zhou Q (2015) Generation of an LncRNA Gtl2-GFP reporter for rapid assessment of pluripotency in mouse induced pluripotent stem cells. J Genet Genomics 42:125–128. doi:10.1016/j.jgg.2015.02.004
Rapicavoli NA, Poth EM, Blackshaw S (2010) The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 10:49. doi:10.1186/1471-213x-10-49
Acknowledgments
This study was supported by grants from the Korean Health Technology R&D Project (HI14C2348), Ministry of Health and Welfare, Republic of Korea, and the National Research Foundation (NRF) of Korea (2011–0012728). S-T.L was supported by the Seoul National University Hospital Research Fund (04-2014-0730) and Korean Health Technology R&D Project (HI12C1773).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding
This study was supported by grants from the Korean Health Technology R&D Project (HI14C2348), Ministry of Health and Welfare, Republic of Korea, and the National Research Foundation (NRF) of Korea (2011–0012728). S-T.L was supported by the Seoul National University Hospital Research Fund (04-2014-0730) and Korean Health Technology R&D Project (HI12C1773).
Additional information
Jun-Sang Sunwoo and Soon-Tae Lee contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 32 kb)
Rights and permissions
About this article
Cite this article
Sunwoo, JS., Lee, ST., Im, W. et al. Altered Expression of the Long Noncoding RNA NEAT1 in Huntington’s Disease. Mol Neurobiol 54, 1577–1586 (2017). https://doi.org/10.1007/s12035-016-9928-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-9928-9