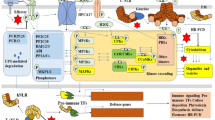
Abstract
Tea, which is mainly produced using the young leaves and buds of tea plants (Camellia sinensis (L.) O. Kuntze), is one of the most common non-alcoholic beverages consumed in the world. The standard of tea mostly depends on the variety and quality of tea plants, which generally grow in subtropical areas, where the warm and humid conditions are also conducive to the occurrence of diseases. In fighting against pathogens, plants rely on their sophisticated innate immune systems which has been extensively studied in model plants. Many components involved in pathogen associated molecular patterns (PAMPs) triggered immunity (PTI) and effector triggered immunity (ETI) have been found. Nevertheless, the molecular regulating network against pathogens (e.g., Pseudopestalotiopsis sp., Colletotrichum sp. and Exobasidium vexans) causing widespread disease (such as grey blight disease, anthracnose, and blister blight) in tea plants is still unclear. With the recent release of the genome data of tea plants, numerous genes involved in tea plant immunity have been identified, and the molecular mechanisms behind tea plant immunity is being studied. Therefore, the recent achievements in identifying and cloning functional genes/gene families, in finding crucial components of tea immunity signaling pathways, and in understanding the role of secondary metabolites have been summarized and the opportunities and challenges in the future studies of tea immunity are highlighted in this review.
Similar content being viewed by others
References
Samanta S (2022) Potential bioactive components and health promotional benefits of tea (Camellia sinensis). J Am Nutr Assoc 41:65–93. https://doi.org/10.1080/07315724.2020.1827082
Unno K, Yamada H, Iguchi K, Ishida H, Iwao Y, Morita A, Nakamura Y (2017) Anti-stress effect of green tea with lowered caffeine on humans: a pilot study. Biol Pharm Bull 40:902–909. https://doi.org/10.1248/bpb.b17-00141
Liu C, Guo YT, Sun LL, Lai XF, Li QH, Zhang WJ, Xiang LM, Sun SL, Cao FR (2019) Six types of tea reduce high-fat-diet-induced fat accumulation in mice by increasing lipid metabolism and suppressing inflammation. Food Funct 10:2061–2074. https://doi.org/10.1039/c8fo02334d
Pan SY, Nie Q, Tai HC, Song XL, Tong YF, Zhang LJF, Wu XW, Lin ZH, Zhang YY, Ye DY, Zhang Y, Wang XY, Zhu PL, Chu ZS, Yu ZL, Liang C (2022) Tea and tea drinking: China’s outstanding contributions to the mankind. Chin Med 17:27. https://doi.org/10.1186/s13020-022-00571-1
Xia EH, Zhang HB, Sheng J, Li K, Zhang QJ, Kim C, Zhang Y, Liu Y, Zhu T, Li W, Huang H, Tong Y, Nan H, Shi C, Shi C, Jiang JJ, Mao SY, Jiao JY, Zhang D, Zhao Y, Zhao YJ, Zhang LP, Liu YL, Liu BY, Yu Y, Shao SF, Ni DJ, Eichler EE, Gao LZ (2017) The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol Plant 10:866–877. https://doi.org/10.1016/j.molp.2017.04.002
Pandey AK, Sinniah GD, Babu A, Tanti A (2021) How the global tea industry copes with fungal diseases—challenges and opportunities. Plant Dis 105:1868–1879. https://doi.org/10.1094/PDIS-09-20-1945-FE
Nisha SN, Prabu G, Mandal AKA (2018) Biochemical and molecular studies on the resistance mechanisms in tea [Camellia sinensis (L.) O. Kuntze] against blister blight disease. Physiol Mol Biol Plants 24:867–880. https://doi.org/10.1007/s12298-018-0565-9
Chen YJ, Zeng L, Shu N, Jiang MY, Wang H, Huang YJ, Tong HR (2018) Pestalotiopsis-like species causing gray blight disease on Camellia sinensis in China. Plant Dis 102:98–106. https://doi.org/10.1094/PDIS-05-17-0642-RE
Wang SS, Mi XZ, Wu ZR, Zhang LX, Wei CL (2019) Characterization and pathogenicity of Pestalotiopsis-like species associated with gray blight disease on Camellia sinensis in Anhui province, China. Plant Dis 103:2786–2797. https://doi.org/10.1094/PDIS-02-19-0412-RE
Kibblewhite MG, Prakash S, Hazarika M, Burgess PJ, Sakrabani R (2014) Managing declining yields from ageing tea plantations. J Sci Food Agric 94:1477–1481. https://doi.org/10.1002/jsfa.6543
Zhang ZB, Feng XB, Wang Y, Xu WW, Huang K, Hu MH, Zhang C, Yuan HY (2019) Advances in research on functional genes of tea plant. Gene 711:143940. https://doi.org/10.1016/j.gene.2019.143940
Wei CL, Yang H, Wang SB, Zhao J, Liu C, Gao LP, Xia EH, Lu Y, Tai YL, She GB, Sun J, Cao HS, Tong W, Gao Q, Li YY, Deng WW, Jiang XL, Wang WZ, Chen Q, Zhang SH, Li HJ, Wu JL, Wang P, Li PH, Shi CY, Zheng FY, Jian JB, Huang B, Shan D, Shi MM, Fang CB, Yue Y, Li FD, Li DX, Wei S, Han B, Jiang CJ, Yin Y, Xia T, Zhang ZZ, Bennetzen JL, Zhao SC, Wan XC (2018) Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci USA 115:E4151–E4158. https://doi.org/10.1073/pnas.1719622115
Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341:746–751. https://doi.org/10.1126/science.1236011
Tang DZ, Wang GX, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29:618–637. https://doi.org/10.1105/tpc.16.00891
Cao YR, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3:e03766. https://doi.org/10.7554/eLife.03766
Gong BQ, Xue J, Zhang NN, Xu LH, Yao XR, Yang QJ, Yu Y, Wang HB, Zhang DD, Li JF (2017) Rice chitin receptor OsCEBiP is not a transmembrane protein but targets the plasma membrane via a GPI anchor. Mol Plant 10:767–770. https://doi.org/10.1016/j.molp.2016.12.005
Gong BQ, Wang FZ, Li JF (2020) Hide-and-seek: chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci 25:805–816. https://doi.org/10.1016/j.tplants.2020.03.006
Chatterjee A, Paul A, Unnati GM, Rajput R, Biswas T, Kar T, Basak S, Mishra N, Pandey A, Srivastava AP (2020) MAPK cascade gene family in Camellia sinensis: in-silico identification, expression profiles and regulatory network analysis. BMC Genomics 21:613. https://doi.org/10.1186/s12864-020-07030-x
Paul A, Srivastava AP, Subrahmanya S, Shen GX, Mishra N (2021) In-silico genome wide analysis of mitogen activated protein kinase kinase kinase gene family in C. sinensis. PLoS One 16:e0258657. https://doi.org/10.1371/journal.pone.0258657
Wang W, Feng BM, Zhou JM, Tang DZ (2020) Plant immune signaling: advancing on two frontiers. J Integr Plant Biol 62:2–24. https://doi.org/10.1111/jipb.12898
Monteiro F, Nishimura MT (2018) Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu Rev Phytopathol 56:243–267. https://doi.org/10.1146/annurev-phyto-080417-045817
Yuan M, Ngou BPM, Ding PT, Xin XF (2021) PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62:102030. https://doi.org/10.1016/j.pbi.2021.102030
Wang YC, Lu QH, Xiong F, Hao XY, Wang L, Zheng MX, Li NN, Ding CQ, Wang XC, Yang YJ (2020) Genome-wide identification, characterization, and expression analysis of nucleotide-binding leucine-rich repeats gene family under environmental stresses in tea (Camellia sinensis). Genomics 112:1351–1362. https://doi.org/10.1016/j.ygeno.2019.08.004
Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, Casey LW, Lonhienne T, Winzor DJ, Zhang XX, Coerdt A, Parker JE, Dodds PN, Kobe B, Jones JDG (2014) Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344:299–303. https://doi.org/10.1126/science.1247357
Shi YL, Sheng YY, Cai ZY, Yang R, Li QS, Li XM, Li D, Guo XY, Lu JL, Ye JH, Wang KR, Zhang LJ, Liang YR, Zheng XQ (2019) Involvement of salicylic acid in anthracnose infection in tea plants revealed by transcriptome profiling. Int J Mol Sci 20:2439. https://doi.org/10.3390/ijms20102439
Lee HI, Leon J, Raskin I (1995) Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci USA 92:4076–4079. https://doi.org/10.1073/pnas.92.10.4076
Camera SL, Gouzerh G, Dhondt S, Hoffmann L, Fritig B, Legrand M, Heitz T (2004) Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol Rev 198:267–284. https://doi.org/10.1111/j.0105-2896.2004.0129.x
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20. https://doi.org/10.1093/mp/ssp106
Huang Jl GuM, Lai ZB, Fan BF, Shi K, Zhou YH, Yu JQ, Chen ZX (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153:1526–1538. https://doi.org/10.1104/pp.110.157370
Tonnessen BW, Manosalva P, Lang JM, Baraoidan M, Bordeos A, Mauleon R, Oard J, Hulbert S, Leung H, Leach JE (2015) Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol Biol 87:273–286. https://doi.org/10.1007/s11103-014-0275-9
Wu YL, Wang WZ, Li YZ, Dai XL, Ma GL, Xing DW, Zhu MQ, Gao LP, Xia T (2017) Six phenylalanine ammonia-lyases from Camellia sinensis: evolution, expression, and kinetics. Plant Physiol and Biochem 118:413–421. https://doi.org/10.1016/j.plaphy.2017.06.030
Wu ZH, Gui ST, Wang SZ, Ding Y (2014) Molecular evolution and functional characterisation of an ancient phenylalanine ammonia-lyase gene (NnPAL1) from Nelumbo nucifera: novel insight into the evolution of the PAL family in Angiosperms. BMC Evol Biol 14:100. https://doi.org/10.1186/1471-2148-14-100
Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y (2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172:47–62. https://doi.org/10.1111/j.1469-8137.2006.01798.x
Vlot AC, Klessig DF, Park SW (2008) Systemic acquired resistance: the elusive signal(s). Curr Opin Plant Biol 11:436–442. https://doi.org/10.1016/j.pbi.2008.05.003
Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9:e0156. https://doi.org/10.1199/tab.0156
Chen Z, Malamy J, Henning J, Conrath U, Sanchez-Casas P, Silva H, Ricigliano J, Klessig DK (1995) Induction, modification, and transduction of the salicylic acid signal in plant defense responses. Proc Natl Acad Sci USA 92:4134–4137. https://doi.org/10.1073/pnas.92.10.4134
Park SW, Kaimoyo E, Kumar D, Mosher S, Klessig DF (2007) Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318:113–116. https://doi.org/10.1126/science.1147113
Ross JR, Nam KH, D’Auria JC, Pichersky E (1999) S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch Biochem Biophys 367:9–16. https://doi.org/10.1006/abbi.1999.1255
Deng WW, Wang RX, Yang TY, Jiang LN, Zhang ZZ (2017) Functional characterization of salicylic acid carboxyl methyltransferase from Camellia sinensis, providing the aroma compound of methyl salicylate during the withering process of white tea. J Agric Food Chem 65:11036–11045. https://doi.org/10.1021/acs.jafc.7b04575
Hu YQ, Zhang MT, Lu MQ, Wu Y, Jing TT, Zhao MY, Zhao YF, Feng YY, Wang JM, Gao T, Zhou ZX, Wu B, Jiang H, Wan XC, Schwab W, Song CK (2022) Salicylic acid carboxyl glucosyltransferase UGT87E7 regulates disease resistance in Camellia sinensis. Plant Physiol 188:1507–1520. https://doi.org/10.1093/plphys/kiab569
Shi CY, Yang H, Wei CL, Yu O, Zhang ZZ, Jiang CJ, Sun J, Li YY, Chen Q, Xia T, Wan XC (2011) Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics 12:131. https://doi.org/10.1186/1471-2164-12-131
Wang L, Wang YC, Cao HL, Hao XY, Zeng JM, Yang YJ, Wang XC (2016) Transcriptome analysis of an anthracnose-resistant tea plant cultivar reveals genes associated with resistance to Colletotrichum camelliae. PLoS ONE 11:e0148535. https://doi.org/10.1371/journal.pone.0148535
Wang SS, Liu L, Mi XZ, Zhao SQ, An YL, Xia XB, Guo R, Wei CL (2021) Multi-omics analysis to visualize the dynamic roles of defense genes in the response of tea plants to gray blight. Plant J 106:862–875. https://doi.org/10.1111/tpj.15203
Wang YC, Hao XY, Lu QH, Wang L, Qian WJ, Li NN, Ding CQ, Wang XC, Yang YJ (2018) Transcriptional analysis and histochemistry reveal that hypersensitive cell death and H2O2 have crucial roles in the resistance of tea plant (Camellia sinensis (L.) O. Kuntze) to anthracnose. Hortic Res 5:18. https://doi.org/10.1038/s41438-018-0025-2
Jeyaraj A, Wang XW, Wang SS, Liu SR, Zhang R, Wu AL, Wei CL (2019) Identification of regulatory networks of MicroRNAs and their targets in response to Colletotrichum gloeosporioides in tea plant (Camellia sinensis L.). Front Plant Sci 10:1096. https://doi.org/10.3389/fpls.2019.01096
Wang SS, Liu SR, Liu L, Li R, Guo R, Xia XB, Wei CL (2020) miR477 targets the phenylalanine ammonia-lyase gene and enhances the susceptibility of the tea plant (Camellia sinensis) to disease during Pseudopestalotiopsis species infection. Planta 251:59. https://doi.org/10.1007/s00425-020-03353-x
Jayaswall K, Mahajan P, Singh G, Parmar R, Seth R, Raina A, Swarnkar MK, Singh AK, Shankar R, Sharma RK (2016) Transcriptome analysis reveals candidate genes involved in blister blight defense in tea (Camellia sinensis (L) Kuntze). Sci Rep 6:30412. https://doi.org/10.1038/srep30412
Zhang QJ, Li W, Li K, Nan H, Shi C, Zhang Y, Dai ZY, Lin YL, Yang XL, Tong Y, Zhang D, Lu C, Feng LY, Wang CF, Liu XX, Huang JA, Jiang WK, Wang XH, Zhang XC, Eichler EE, Liu ZH, Gao LZ (2020) The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution. Mol Plant 13:935–938. https://doi.org/10.1016/j.molp.2020.04.009
Wang XC, Feng H, Chang YX, Ma CL, Wang LY, Hao XY, Li AL, Cheng H, Wang L, Cui P, Jin JQ, Wang XB, Wei K, Ai C, Zhao S, Wu ZC, Li YY, Liu BY, Wang GD, Chen L, Ruan J, Yang YJ (2020) Population sequencing enhances understanding of tea plant evolution. Nat Commun 11:4447. https://doi.org/10.1038/s41467-020-18228-8
Zhang XT, Chen S, Shi LQ, Gong DP, Zhang SC, Zhao Q, Zhan DL, Vasseur L, Wang YB, Yu JX, Liao ZY, Xu XD, Qi R, Wang WL, Ma YR, Wang PJ, Ye NX, Ma DN, Shi Y, Wang HF, Ma XK, Kong XR, Lin J, Wei LF, Ma YY, Li RY, Hu GP, He HF, Zhang L, Ming R, Wang G, Tang HB, You MS (2021) Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat Genet 53:1250–1259. https://doi.org/10.1038/s41588-021-00895-y
Wang PJ, Yue C, Chen D, Zheng YC, Zhang Q, Yang JF, Ye NX (2019) Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes Genomics 41:17–33. https://doi.org/10.1007/s13258-018-0734-9
Liu RJ, Wang YY, Tang S, Cai JR, Liu SQ, Zheng P, Sun BM (2021) Genome-wide identification of the tea plant bHLH transcription factor family and discovery of candidate regulators of trichome formation. Sci Rep 11:10764. https://doi.org/10.1038/s41598-021-90205-7
Zhang ZB, Jin YJ, Wan HH, Cheng L, Feng ZG (2021) Genome-wide identification and expression analysis of the MADS-box transcription factor family in Camellia sinensis. J Appl Genet 62:249–264. https://doi.org/10.1007/s13353-021-00621-8
Wang YX, Liu ZW, Wu ZJ, Li H, Wang WL, Cui X, Zhuang J (2018) Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant (Camellia sinensis). Sci Rep 8:3949. https://doi.org/10.1038/s41598-018-22275-z
Zheng YC, Chen XJ, Wang PJ, Sun Y, Yue C, Ye NX (2020) Genome-wide and expression pattern analysis of JAZ family involved in stress responses and postharvest processing treatments in Camellia sinensis. Sci Rep 10:2792. https://doi.org/10.1038/s41598-020-59675-z
Chen JF, Gao T, Wan SQ, Zhang YH, Yang JK, Yu YB, Wang WD (2018) Genome-wide identification, classification and expression analysis of the HSP gene superfamily in tea plant (Camellia sinensis). Int J Mol Sci 19:2633. https://doi.org/10.3390/ijms19092633
Bordoloi KS, Krishnatreya DB, Baruah PM, Borah AK, Mondal TK, Agarwala N (2021) Genome-wide identification and expression profiling of chitinase genes in tea (Camellia sinensis (L.) O. Kuntze) under biotic stress conditions. Physiol Mol Biol Plants 27:369–385. https://doi.org/10.1007/s12298-021-00947-x
Zhang QQ, Guo NN, Zhang YH, Yu YB, Liu SY (2022) Genome-wide characterization and expression analysis of Pathogenesis-Related 1 (PR-1) gene Family in Tea Plant (Camellia sinensis (L.) O. Kuntze) in response to blister-blight disease Stress. Int J Mol Sci 23:1292. https://doi.org/10.3390/ijms23031292
Jesus-Pires CD, Ferreira-Neto JRC, Bezerra-Neto JP, Kido EA, Silva RLDO, Pandolfi V, Wanderley-Nogueira AC, Binneck E, Costa AFD, Pio-Ribeiro G, Pereira-Andrade G, Sittolin IM, Freire-Filho F, Benko-Iseppon AM (2020) Plant Thaumatin-like proteins: function, evolution and biotechnological applications. Curr Protein Pept Sci 21:36–51. https://doi.org/10.2174/1389203720666190318164905
Acharya K, Pal AK, Gulati A, Kumar S, Singh AK, Ahuja PS (2013) Overexpression of Camellia sinensis Thaumatin-like protein, CsTLP in potato confers enhanced resistance to Macrophomina phaseolina and Phytophthora infestans infection. Mol Biotechnol 54:609–622. https://doi.org/10.1007/s12033-012-9603-y
Rustgi S, Boex-Fontvieille E, Reinbothe C, Wettstein DV, Reinbot S (2018) The complex world of plant protease inhibitors: insights into a Kunitz-type cysteine protease inhibitor of Arabidopsis thaliana. Commun Integr Biol 11:e1368599. https://doi.org/10.1080/19420889.2017.1368599
Boex-Fontvieille E, Rustgi S, Reinbothe S, Reinbothe C (2015) A Kunitz-type protease inhibitor regulates programmed cell death during flower development in Arabidopsis thaliana. J Exp Bot 66:6119–6135. https://doi.org/10.1093/jxb/erv327
Boex-Fontvieille E, Rustgi S, Wettstein DV, Reinbothe S, Reinbothe C (2015) Water-soluble chlorophyll protein is involved in herbivore resistance activation during greening of Arabidopsis thaliana. Proc Natl Acad Sci USA 112:7303–7308. https://doi.org/10.1073/pnas.1507714112
Budic M, Sabotic J, Meglic V, Kos J, Kidric M (2013) Characterization of two novel subtilases from common bean (Phaseolus vulgaris L.) and their responses to drought. Plant Physiol Biochem 62:79–87. https://doi.org/10.1016/j.plaphy.2012.10.022
Zhu JY, He YX, Yan XM, Liu L, Guo R, Xia XB, Cheng DJ, Mi XZ, Samarina L, Liu SR, Xia EH, Wei CL (2019) Duplication and transcriptional divergence of three Kunitz protease inhibitor genes that modulate insect and pathogen defenses in tea plant (Camellia sinensis). Hortic Res 6:126. https://doi.org/10.1038/s41438-019-0208-5
Tibpromma S, Dong Y, Ranjitkar S, Schaefer DA, Karunarathna SC, Hyde KD, Jayawardena RS, Manawasinghe IS, Bebber DP, Promputtha I, Xu JC, Mortimer PE, Sheng J (2021) Climate-fungal pathogen modeling predicts loss of up to one-third of tea growing areas. Front Cell Infect Microbiol 11:610567. https://doi.org/10.3389/fcimb.2021.610567
Zhang S, Li X, Sun ZH, Shao SJ, Hu LF, Ye M, Zhou YH, Xia XJ, Yu JQ, Shi K (2015) Antagonism between phytohormone signalling underlies the variation in disease susceptibility of tomato plants under elevated CO2. J Exp Bot 66:1951–1963. https://doi.org/10.1093/jxb/eru538
Sowndhararajan K, Marimuthu S, Manian S (2013) Integrated control of blister blight disease in tea using the biocontrol agent Ochrobactrum anthropi strain BMO-111 with chemical fungicides. J Appl Microbiol 114:1491–1499. https://doi.org/10.1111/jam.12159
Borah A, Hazarika SN, Thakur D (2022) Potentiality of actinobacteria to combat against biotic and abiotic stresses in tea [Camellia sinensis (L) O. Kuntze]. J Appl Microbiol 133:2314–2330. https://doi.org/10.1111/jam.15734
Wang YC, Qian WJ, Li NN, Hao XY, Wang L, Xiao B, Wang XC, Yang YJ (2016) Metabolic changes of caffeine in tea plant (Camellia sinensis (L.) O. Kuntze) as defense response to Colletotrichum fructicola. J Agric Food Chem 64:6685–6693. https://doi.org/10.1021/acs.jafc.6b02044
Chen SL, Zhang LP, Cai XM, Li X, Bian L, Luo ZX, Li ZQ, Chen ZM, Xin ZJ (2020) (E)-Nerolidol is a volatile signal that induces defenses against insects and pathogens in tea plants. Hortic Res 7:52. https://doi.org/10.1038/s41438-020-0275-7
Kim YS, Sano H (2008) Pathogen resistance of transgenic tobacco plants producing caffeine. Phytochemistry 69:882–888. https://doi.org/10.1016/j.phytochem.2007.10.021
Li X, Ahammed GJ, Li ZX, Tang MJ, Yan P, Han WY (2016) Decreased biosynthesis of jasmonic acid via lipoxygenase pathway compromised caffeine-induced resistance to Colletotrichum gloeosporioides under elevated CO2 in tea seedlings. Phytopathology 106:1270–1277. https://doi.org/10.1094/PHYTO-12-15-0336-R
Acknowledgements
The authors sincerely acknowledge Dr. Qing Xiong (Sichuan Agricultural University, Chengdu, China) for critical comments and linguistic assistance during the preparation of this manuscript. We also thank Dr. Xia Yan (Chongqing Three Gorges University, Wanzhou, China) for her contribution in improving the manuscript.
Funding
This work was supported by the Chongqing Natural Science Foundation Project (cstc2021jcyj-msxmX0322), and the Science and Technology Project of Chongqing Municipal Education Commission (KJQN202101246).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. The first draft of the manuscript was written by LR, JS, TL, JS and all authors commented on previous versions of the manuscript. The manuscript was revised by LR. LR and GW approved the final version of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest is reported by the authors.
Research involving human and animals rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rui, L., Su, Jy., Li, T. et al. Molecular regulation of immunity in tea plants. Mol Biol Rep 50, 2883–2892 (2023). https://doi.org/10.1007/s11033-022-08177-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08177-4