Abstract
Background
Methemoglobin is the reduced form of haemoglobin that is normally found in the blood in levels < 1%. Methemoglobinemia can occur as a congenital or acquired disease. Two types of recessive congenital methaemoglobinemia (RCM) are caused by the NADH-dependent cytochrome b5 reductase enzyme deficiency of the CYB5R3 gene. RCM-I is characterized by higher methaemoglobin levels (> 2 g/dL), causing only cyanosis, whereas RCM-II is associated with cyanosis with neurological impairment.
Methods
Routine haematological investigations were done by standard method. The methaemoglobin level was evaluated by the potassium ferricyanide assay. NADH-cytochrome b5 reductase (cytb5r) enzyme activities were measured by standard methods, and molecular analysis was performed by polymerase chain reaction (PCR) followed by DNA sequencing. The interpretation of mutation effect and the molecular modeling were performed by using specific software DEEP VIEW SWISS-PDB VIEWER and Pymol molecular graphics program.
Results
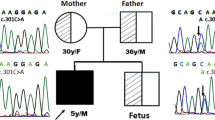
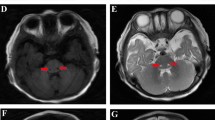
The present study discovered three novel homozygous pathogenic variants of CYB5R3 causing RCM I and II in four unrelated Indian patients. In patient-1 and patient-2 of RCM type I caused due to novel c.175C>T (p.Arg59Cys) and other reported c.469T>C (p.Phe157Ser) missense pathogenic variants respectively, whereas patient-3 and patient-4 presented with the RCM type II are related to developmental delay with cyanosis since birth due to a novel homozygous (g.25679_25679delA) splice-site deletion and novel homozygous c.824_825insC (p.Pro278ThrfsTer367) single nucleotide insertion. The CYB5R3 transcript levels were estimated by qRT-PCR in the splice-site deletion, which was 0.33fold of normal healthy control. The insertion of nucleotide C resulted in a frameshift of termination codon are associated with neurological impairment.
Conclusions
Molecular diagnosis of RCM can help to conduct genetic counselling for novel mutations and, subsequently, prenatal diagnosis of high-risk genetic disorders.


Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
References
Scott EM, Griffith IV (1959) The enzymic defect of hereditary methemoglobinemia: diaphorase. BBA Biochim Biophys Acta 34:584–586. https://doi.org/10.1016/0006-3002(59)90324-5
Rehman HU (2001) Methemoglobinemia. West J Med 175:193–196
Elahian F, Sepehrizadeh Z, Moghimi B, Mirzaei SA (2014) Human cytochrome b5 reductase: Structure, function, and potential applications. Crit Rev Biotechnol 34:134–143. https://doi.org/10.3109/07388551.2012.732031
Skold A, Cosco DL, Klein R (2011) Methemoglobinemia. South Med J 104:757–761. https://doi.org/10.1097/SMJ.0b013e318232139f
Fukushima H, Grinstead GF, Gaylor JL (1981) Total enzymic synthesis of cholesterol from lanosterol. Cytochrome b5-dependence of 4-methyl sterol oxidase. J Biol Chem 256:4822–4826
Siendones E, Ballesteros M, Navas P (2018) Cellular and molecular mechanisms of recessive hereditary methaemoglobinaemia type II. J Clin Med 7:341. https://doi.org/10.3390/jcm7100341
Junien C, Vibert M, Weil D, Van-Cong N, Kaplan J-C (1978) Assignment of NADH-cytochrome b5 reductase (DIA1 locus) to human chromosome 22. Hum Genet 42:233–239. https://doi.org/10.1007/BF00291301
Choury D, Leroux A, Kaplan JC (1981) Membrane-bound cytochrome b5 reductase (methemoglobin reductase) in human erythrocytes. Study in normal and methemoglobinemic subjects. J Clin Invest 67:149–155. https://doi.org/10.1172/JCI110007
Fisher RA, Povey S, Bobrow M, Solomon E, Boyd Y, Carritt B (1977) Assignment of the DIA1 locus to chromosome 22. Ann Hum Genet 41:151–155. https://doi.org/10.1111/j.1469-1809.1977.tb01909.x
Siendones E, Santa C-C, Martin-Montalvo A, Cascajo MV, Ariza J, Lopez-Lluch G, Villalba JM, Acquaviva-Bourdain C, Roze E, Bernier M et al (2014) Membrane-bound CYB5R3 is a common effector of nutritional and oxidative stress response through FOXO3a and Nrf2. Antioxid Redox Signal 21:1708–1725. https://doi.org/10.1089/ars.2013.5479
Borgese N, D’Arrigo A, De Silvestris M, Pietrini G (1993) NADH-cytochrome b5 reductase and cytochrome b5. The problem of posttranslational targeting to the endoplasmic reticulum. Subcell Biochem 21:313–341
Leroux A, Mota VL, Kahn A (2001) Transcriptional and translational mechanisms of cytochrome b5 reductase isoenzyme generation in humans. Biochem J 355:529–535. https://doi.org/10.1042/bj3550529
Scott EM, Griffith IV (1959) The enzymic defect of hereditary methemoglobinemia: diaphorase. Biochim Biophys Acta 34:584–586. https://doi.org/10.1016/0006-3002(59)90324-5
Hultquist DE, Passon PG (1971) Catalysis of methaemoglobin reduction by erythrocyte cytochrome b5 and cytochrome b5 reductase. Nature 229:252–254. https://doi.org/10.1038/newbio229252a0
Percy MJ, Lappin TR (2008) Recessive congenital methaemoglobinemia: cytochrome b5 reductase deficiency. Br J Haematol 141:298–308
Ewenczyk C, Leroux A, Roubergue A, Laugel V, Afenjar A, Saudubray JM, Beauvais P, de Villemeur TB, Vidailhet M, Roze E (2008) Recessive hereditary methaemoglobinaemia, type II: delineation of the clinical spectrum. Brain 131:760–761. https://doi.org/10.1093/brain/awm337
Gupta V, Kulkarni A, Warang P, Devendra R, Chiddarwar A, Kedar P (2020) Mutation update: variants of the CYB5R3 gene in recessive congenital methemoglobinemia. Hum Mutat 41:737–748. https://doi.org/10.1002/humu.23973
Lewis SM, Barbara JB, Bates I (2006) Dacie and Lewis practical haematology, 11th edn. Elsevier, Amsterdam. https://doi.org/10.1016/B0-443-06660-4/X5001-6
Bain BJ, Bates I, Laffan MA, Lewis SM (2011) Basic hematological techniques. Springer, New York
Evelyn KA, Malloy HT (1938) Microdetermination of oxyhemoglobin, methemoglobin, and sulfhemoglobin in a single sample of blood. J Biol Chem 126:655–662
Fujii S, Dale GL, Beutler E (1984) Glutathione-dependent protection against oxidative damage of the human red cell membrane. Blood 63:1096–1101. https://doi.org/10.1182/blood.v63.5.1096.bloodjournal6351096
Mannino EA, Pluim T, Wessler J, Cho MT, Juusola J, Schrier Vergano SA (2018) Congenital methemoglobinemia type II in a 5-year-old boy. Clin Case Rep 6:170–178. https://doi.org/10.1002/ccr3.1310
Grantham R (1974) Amino acid difference formula to help explain protein evolution. Science 185:862–864. https://doi.org/10.1126/science.185.4154.862
Pandurangan AP, Ochoa-Montaño B, Ascher DB, Blundell TL (2017) SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res 45:W229–W235. https://doi.org/10.1093/nar/gkx439
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. https://doi.org/10.1093/bioinformatics/btv195
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using polyphen-2. Curr Protoc Hum Genet 76:20.1-20.41. https://doi.org/10.1002/0471142905.hg0720s76
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31:3812–3814. https://doi.org/10.1093/nar/gkg509
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. https://doi.org/10.1093/nar/gkv342
Brogna S, Wen J (2009) Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol 16:107–113
Bittles A (2001) Consanguinity and its relevance to clinical genetics. Clin Genet 60:89–98. https://doi.org/10.1034/j.1399-0004.2001.600201.x
Acknowledgements
We would like to thank patients and family members for their cooperation and participation in this study. This study was performed with the Indian Council of Medical Research New Delhi and the Council of Scientific and Industrial Research, India (CSIR) New Delhi for financial support.
Funding
This study was performed with the financial support of the Indian Council of Medical Research New Delhi and also supported by the Council of Scientific and Industrial Research (CSIR), India, Grant No. 27(0333)/18/EMR-II).
Author information
Authors and Affiliations
Contributions
PK: Conceived ideas and initiated the experimental work and supervision of work. AK: Contributed to the findings of the work. AD: Further experimental work and statistical analysis for expansion of idea were carried under the supervision of PK, AD, and PK: Manuscript was written by AD in consultation with PK.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interests.
Ethical approval
This study was approved by the Ethics Committee of the Institutional Ethical Committee of ICMR-National Institute of Immunohaematology, Mumbai. All procedures performed in studies involving human participants were as per the ethical standards of the institutional review board of the Institute and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent to participate
Written informed consent was obtained from a legally authorized representative(s) for anonymized patient information to be published in this article.
Consent for publication
The written informed consent for publication was obtained.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Deorukhkar, A., Kulkarni, A. & Kedar, P. Three novel mutations in CYB5R3 gene causing NADH-cytochrome b5 reductase enzyme deficiency leads to recessive congenital methaemoglobinemia. Mol Biol Rep 49, 2141–2147 (2022). https://doi.org/10.1007/s11033-021-07031-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-07031-3