Abstract
Background
As one of the most important factors of the japonica rice plant, leaf shape affects the photosynthesis and carbohydrate accumulation directly. Mining and using new leaf shape related genes/QTLs can further enrich the theory of molecular breeding and accelerate the breeding process of japonica rice.
Methods
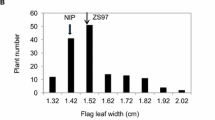
In the present study, 2 RILs and a natural population with 295 japonica rice varieties were used to map QTLs for flag leaf length (FL), flag leaf width (FW) and flag leaf area (FLA) by linkage analysis and genome-wide association study (GWAS) throughout 2 years.
Results
A total of 64 QTLs were detected by 2 ways, and pleiotropic QTLs qFL2 (Chr2_33,332,579) and qFL10 (Chr10_10,107,835; Chr10_10,230,100) consisted of overlapping QTLs mapped by linkage analysis and GWAS throughout the 2 years were identified.
Conclusions
The candidate genes LOC_Os02g54254, LOC_Os02g54550, LOC_Os10g20160, LOC_Os10g20240, LOC_Os10g20260 were obtained, filtered by linkage disequilibrium (LD), and haplotype analysis. LOC_Os10g20160 (SD-RLK-45) showed outstanding characteristics in quantitative real-time PCR (qRT-PCR) analysis in leaf development period, belongs to S-domain receptor-like protein kinases gene and probably to be a main gene regulating flag leaf width of japonica rice. The results of this study provide valuable resources for mining the main genes/QTLs of japonica rice leaf development and molecular breeding of japonica rice ideal leaf shape.



Similar content being viewed by others
References
Pane TC, Supriyono Y, Novita D (2021) Supporting food security with rice farming insurance: the farmers’ perceptions (case study in cinta damai village, percut sei tuan subdistrict, deli serdang district). IOP Conf Ser 782(2):022044. https://doi.org/10.1088/1755-1315/782/2/022044
Barboza LGA, Vethaak AD, Lavorante BR, Lundebye AK, Guilhermino L (2018) Marine microplastic debris: an emerging issue for food security, food safety and human health. Marine Pollut Bull 133:336–348. https://doi.org/10.1016/j.marpolbul.2018.05.047
Timmer CP (2014) Food security in Asia and the pacific: the rapidly changing role of rice. Asia Pacific Policy Stud. https://doi.org/10.1002/app5.6
Adachi M, Hasegawa T, Fukayama H, Tokida T, Sakai H, Matsunami T, Nakamura H, Sameshima R, Okada M et al (2014) Soil and water warming accelerates phenology and down-regulation of leaf photosynthesis of rice plants grown under free-air co2 enrichment (face). Plant Cell Physiol. https://doi.org/10.1093/pcp/pcu005
Zhu C, Zhu J, Cao J, Jiang Q, Liu G, Ziska LH (2014) Biochemical and molecular characteristics of leaf photosynthesis and relative seed yield of two contrasting rice cultivars in response to elevated [CO2]. J Exp Bot 65(20):6049. https://doi.org/10.1093/jxb/eru344
Gu J, Yin X, Tjeerd-Jan S, Wang H, Struik PC (2012) Physiological basis of genetic variation in leaf photosynthesis among rice (oryza sativa l.) introgression lines under drought and well-watered conditions. J Exp Bot 14:5137. https://doi.org/10.1093/jxb/ers170
He PL, Wang XW, Zhang XB, Jiang YD, Tian WJ, Zhang XQ, Li YY, Sun Y, Xie J, Ni JL, He GH, Sang XC (2018) Short and narrow flag leaf1, a GATA zinc finger domain-containing protein, regulates flag leaf size in rice (Oryza sativa L.). BMC Plant Biol. https://doi.org/10.1186/s12870-018-1452-9
Rahman MA, Haque ME, Sikdar B, Islam MA, Matin MN (2013) Correlation analysis of flag leaf with yield in several rice cultivars. J Life Earth Sci 8:49–54. https://doi.org/10.3329/jles.v8i0.20139
Giuliani R, Koteyeva N, Voznesenskaya E, Evans MA, Cousins AB, Edwards GE (2013) Coordination of leaf photosynthesis, transpiration, and structural traits in rice and wild relatives (Genus Oryza). Plant Physiol 162:1632–1651. https://doi.org/10.1104/pp.113.217497
Zhu X, Song Q, Ort DR (2012) Elements of a dynamic systems model of canopy photosynthesis. Curr Opin Plant Biol 15:237–244. https://doi.org/10.1016/j.pbi.2012.01.010
Hoang GT, Gantet P, Nguyen KH, Phung N, Xuan HP (2019) Genome-wide association mapping of leaf mass traits in a vietnamese rice landrace panel. PLoS ONE 14(7):e0219274. https://doi.org/10.1371/journal.pone.0219274
Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije M, Sekiguchi H (2008) Narrow leaf 7 controls leaf shape mediated by auxin in rice. Mol Genet Genom 279:499–507. https://doi.org/10.1007/s00438-008-0328-3
Farooq M, Tagle AG, Santos RE, Ebron LA, Fujita D, Kobayashi N (2010) Quantitative trait loci mapping for leaf length and leaf width in rice cv. IR64 derived lines. J Integr Plant Biol 52:578–584. https://doi.org/10.1111/j.1744-7909.2010.00955.x
Chen M, Luo J, Shao G, Wei X, Tang S, Sheng Z, Song J, Hu P (2012) Fine mapping of a major QTL for fag leaf width in rice, qFLW4, which might be caused by alternative splicing of NAL1. Plant Cell Rep 31(863–872):1. https://doi.org/10.1007/s00299-011-1207-7
Zhang G, Li S, Wang L, Ye W, Zeng D, Rao Y, Peng Y, Hu J, Yang Y, Xu J, Ren D, Gao Z, Zhu L, Dong G, Hu X, Yan M, Guo L, Li C, Qian Q (2014) LSCHL4 from Japonica cultivar, which is allelic to NAL1, increases yield of Indica super rice. Mol Plant 7:1350–1364. https://doi.org/10.1093/mp/ssu055
Tang X, Gong R, Sun W et al (2018) Genetic dissection and validation of candidate genes for flag leaf size in rice (Oryza Sativa L.). Theor Appl Genet. https://doi.org/10.1007/s00122-017-3036-8
Zhang B, Ye W, Ren D, Tian P, Peng Y, Gao Y, Ruan B, Wang L, Zhang G, Guo L, Qian Q, Gao Z (2015) Genetic analysis of flag leaf size and candidate genes determination of a major QTL for flag leaf width in rice. Rice (New York, NY) 8(1):39. https://doi.org/10.1186/s12284-014-0039-9
Yang W, Guo Z, Huang C, Wang K, Jiang N, Feng H, Chen G, Liu Q, Xiong L (2015) Genome-wide association study of rice (Oryza sativa L.) leaf traits with a high-throughput leaf scorer. J Exp Bot 66:5605–5615. https://doi.org/10.1093/jxb/erv100
Jiang D, Fang JJ, Lou LM, Zhao JF, Yuan SJ, Yin L, Sun W, Peng LX, Guo BT, Li XY (2015) Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE 10(2):e0118169. https://doi.org/10.1371/journal.pone.0118169
Li A, Yz B, Fang LB, Qian CB, Jq A (2019) Narrow leaf 1 (NAL1) regulates leaf shape by affecting cell expansion in rice (oryza sativa l). Biochem Biophys Res Commun 516(3):957–962. https://doi.org/10.1016/j.bbrc.2019.06.142
Xu J, Wang L, Wang YX, Zeng DL, Zhou MY, Fu X, Ye WJ, Hu J, Zhu L, Ren DY, Gao ZY, Dong GJ, Guo LB, Zhang GH, Qian Q (2017) Reduction of OsFLW7 expression enhanced leaf area and grain production in rice expression enhanced leaf area and grain production in rice. Sci Bull 62(24):1631–1633. https://doi.org/10.1016/j.scib.2017.11.013
Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C et al (2011) Genetic architecture of aluminum tolerance in rice (oryza sativa) determined through genome-wide association analysis and qtl mapping. PLoS Genet 7(8):747–757. https://doi.org/10.1371/journal.pgen.1002221
Li N, Zheng H, Cui J, Wang J, Zou D (2019) Genome-wide association study and candidate gene analysis of alkalinity tolerance in japonica rice germplasm at the seedling stage. Rice. https://doi.org/10.1186/s12284-019-0285-y
Li X, Zheng H, Wu W, Liu H, Zhao H (2020) QTL mapping and candidate gene analysis for alkali tolerance in japonica rice at the bud stage based on linkage mapping and genome-wide association study. Rice. https://doi.org/10.1186/s12284-020-00412-5
Meng L, Li H, Zhang L, Wang J (2015) Qtl icimapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations-sciencedirect. Crop J 3(3):269–283
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Turner SD (2014) Qqman: an r package for visualizing gwas results using q-q and manhattan plots. Biorxiv. https://doi.org/10.1101/005165
Julio R, Albert FM, Sánchez-DelBarrio JC, Sara GR, Pablo L, Ramos-Onsins SE et al (2017) Dnasp 6: dna sequence polymorphism analysis of large data sets. Mol Biol Evol 12:12. https://doi.org/10.1093/molbev/msx248
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001
Aya K, Suzuki G, Suwabe K, Hobo T, Takahashi H, Shiono K et al (2011) Comprehensive network analysis of anther-expressed genes in rice by the combination of 33 laser microdissection and 143 spatiotemporal microarrays. PLoS ONE 6(10):e26162. https://doi.org/10.1371/journal.pone.0026162
Dastan S, Ghareyazie B, Silva J (2020) Selection of ideotype to increase yield potential of gm and non-gm rice cultivars. Plant Sci 279:110519. https://doi.org/10.1016/j.plantsci.2020.110519
Ya-Jie HU, Cao WW, Qian HJ, Xing ZP, Zhang HC, Dai QG et al (2015) Effect of planting density of mechanically transplanted pot seedlings on yield, plant type and lodging resistance in rice with different panicle types. Acta Agron Sin. https://doi.org/10.3724/SP.J.1006.2015.00743
Clerget B, Domingo AJ, Layaoen HL et al (2016) Leaf emergence, tillering, plant growth, and yield in response to plant density in a high-yielding aerobic rice crop. Field Crop Res 199:52–64. https://doi.org/10.1016/j.fcr.2016.09.018
Jiang SK, Zhang XJ, Zheng-Jin XU, Wen-Fu C (2010) Comparison between QTLs for chlorophyll content and genes controlling chlorophyll biosynthesis and degradation in japonica rice. Acta Agron Sin 36(3):376–384. https://doi.org/10.1016/S1875-2780(09)60036-5
Hu W, Zhang H, Jiang J, Wang Y, Sun D, Wang X, Hong D (2012) Discovery of a germplasm with large flag leaf angle and its genetic analysis as well as QTL mapping in japonica rice. Chin J Rice Sci 26(1):34–42
Shen B, Yu WD, Du JH et al (2011) Validation and dissection of quantitative trait loci for leaf traits in interval RM4923-RM402 on the short arm of rice chromosome. Genet 90(1):39–44. https://doi.org/10.1007/s12041-011-0019-4
Zhang B, Ye W, Ren D, Tian P, Peng Y, Gao Y et al (2015) Genetic analysis of flag leaf size and candidate genes determination of a major QTL for flag leaf width in rice. Rice. https://doi.org/10.1186/s12284-014-0039-9
Li D, Wang L, Wang M, Xu Y, Luo W, Liu Y et al (2009) Engineering osbak1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7(8):791–806. https://doi.org/10.1111/j.1467-7652.2009.00444.x
He P, Wang X, Zhang X, Jiang Y, Tian W, Zhang X (2018) Short and narrow flag leaf1, a GATA zinc finger domain-containing protein, regulates flag leaf size in rice (oryza sativa). BMC Plant Biol. https://doi.org/10.1186/s12870-018-1452-9
Zang G, Zou H, Zhang Y, Xiang Z, Huang J, Luo L et al (2016) OsDET1 modulates the aba signaling pathway and aba biosynthesis in rice. Plant Physiol. https://doi.org/10.1104/pp.16.00059
Takaiwa F (2010) Differences in transcriptional regulatory mechanisms functioning for free lysine content and seed storage protein accumulation in rice grain. Plant Cell Physiol 51(12):1964–1974. https://doi.org/10.1093/pcp/pcq164
Wang F, Zhu C (2011) Heterologous expression of a rice syntaxin-related protein KNOLLE gene (OsKNOLLE) in yeast and its functional analysis in the role of abiotic stress. Genetics 33(11):1251–1257
Julian G, Schwerdt K, MacKenzi F et al (2015) Evolutionary dynamics of the cellulose synthase gene superfamily in grasses. Plant Physiol. https://doi.org/10.1104/pp.15.00140
Zou X, Qin Z, Zhang C, Liu B, Liu J, Zhang C et al (2015) Over-expression of an s-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J Exp Bot 22:7197–7209. https://doi.org/10.1093/jxb/erv417
Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol 150(1):12–26. https://doi.org/10.1104/pp.108.134353
Herder GD, Yoshida S, Antolin-Llovera M, Ried MK, Parniske M (2012) Lotus japonicus e3 ligase seven in absentia4 destabilizes the symbiosis receptor-like kinase symrk and negatively regulates rhizobial infection. Plant Cell 24(4):1691–1707. https://doi.org/10.1105/tpc.110.082248
Pan J, Li Z, Wang Q, Yang L, Yao F, Liu W (2019) An s-domain receptor-like kinase, osesg1, regulates early crown root development and drought resistance in rice. Plant Sci. https://doi.org/10.1016/j.plantsci.2019.110318
Morillo SA, Tax FE (2006) Functional analysis of receptor-like kinases in monocots and dicots. Curr Opin Plant Biol 9(5):460–469. https://doi.org/10.1016/j.pbi.2006.07.009
Jinjun Z, Peina JU, Zhang ZH et al (2020) Ossrk1, an atypical s-receptor-like kinase positively regulates leaf width and salt tolerance in rice. Rice Sci 27(2):57–66
Funding
This research was supported by the Heilongjiang Provincial governmental Postdoctoral Foundation of China (LBH-Z16188), the Natural Science Foundation Joint Guide Project of Heilongjiang (LH2019C035) and the Province-Academy Science and Technology Cooperation Project of Heilongjiang (YS20B05). Application R & D Project of Heilongjiang Academy of Agricultural Science (2021YYYF037).
Author information
Authors and Affiliations
Contributions
JW and TW were the main writer of the whole paper. QW, XT, YR, HZ, KL responsible for field data collection. LY mainly engaged in data statistics and data processing. HJ responsible for sequencing data sorting and analysis. YL, QL proofread the full text. DZ, HZ guided the technical route of this study.
Corresponding author
Ethics declarations
Ethical approval
The authors of this paper declare that we have no conflict of interest. Molecular Biology Reports is the only journal we submitted. The submitted work is original and haven’t been published elsewhere in any form or language. This article does not contain any studies with animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, J., Wang, T., Wang, Q. et al. QTL mapping and candidate gene mining of flag leaf size traits in Japonica rice based on linkage mapping and genome-wide association study. Mol Biol Rep 49, 63–71 (2022). https://doi.org/10.1007/s11033-021-06842-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06842-8