Abstract
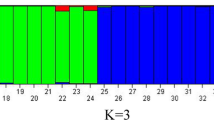
Clerodendrum belonging to the family of Lamiaceae is used in indigenous systems of medicine to treat various life-threatening diseases. The genus has complex morphological variations which lead to limits in its precise taxonomic classifications. Genetic diversity study could enhance taxonomic authentication and evolutionary relationship among the species of Clerodendrum. In this study, nine species of Clerodendrum collected from different regions of North East India were screened using ISSR, RAPD, and SCoT molecular markers. The markers of ISSR, RAPD, and SCoT generated a total of 79, 126, and 145 amplicons with an average of 6.58, 7.86, and 8.53 amplicon per primer. The polymorphism information contents (PIC) for ISSR, RAPD, and SCoT ranged from 0.28 to 0.37, 0.39 to 0.69, and 0.30 to 0.62 with resolving power (Rp) varying from 5.26 to 11.11, 4.04 to 9.67, and 4.54 to 8.65, respectively. Unweighted Pair Group Method with Arithmetic Mean (UPGMA) based clustering methods grouped 94 genotypes into 6 clusters for ISSR and 3 clusters each for RAPD and SCoT markers. Similarly, population structure-based analysis divided 94 genotypes into 6 populations for ISSR and RAPD and 4 populations for SCoT markers. AMOVA analysis revealed that SCoT markers generated maximum genetic variations within and among genotypes, contrary to ISSR and RAPD markers. Results in this study, suggest that the competence of three markers was relatively the same in genotypes fingerprinting, but SCoT was more efficient in the detection of polymorphism for Clerodendrum species. Further, these results could be integrated in the exploration of diverse Clerodendrum species and germplasm utilization.



Similar content being viewed by others
References
Deori C, Roy DK, Talukdar SR, Pagag K, Sarma N (2013) Diversity of the genus Clerodendrum Linnaeus (Lamiaceae) in Northeast India with special reference to Barnadi Wildlife Sanctuary. Assam Pleione 7(2):473–488
Hu HJ, Liu Q, Yang YB, Yang L, Wang ZT (2014) Chemical constituents of Clerodendrum trichotomum leaves. Zhong Yao Cai 37:1590–1593
Steane DA, Scotland RW, Mabberley DJ, Olmstead RG (1999) Molecular systematics of Clerodendrum (Lamiaceae): ITS sequences and total evidence. Am J Bot 86:98–107
Yuan YW, Mabberley DJ, Steane DA, Olmstead RG (2010) Further disintegration and redefinition of Clerodendrum (Lamiaceae): Implications for the understanding of the evolution of an intriguing breeding strategy. Taxon 59(1):125–133
Shrivastava N, Patel T (2007) Clerodendrum and Healthcare: An Overview. Med Aromat Plant Sci Biotechnol 1(1):142–150
Yadav A, Verma PK, Chand T, Bora HR (2018) Ethno-medicinal knowledge of Clerodendrum L. among different tribes of Nambor reserve forest, Assam, India. J. Pharmacogn. Phytochem. 7(5):1567–1570
Yonggam D (2005) Ethno Medico-Botany on the Mishing tribe of East Siang district of Arunachal Pradesh. Arun Forest News 21:44–49
Tiwari G, Singh R, Singh N, Choudhury DR, Paliwal R, Kumar A, Gupta V (2016) Study of arbitrarily amplified (RAPD and ISSR) and gene targeted (SCoT and CBDP) markers for genetic diversity and population structure in Kalmegh [Andrographis paniculata (Burm. f.) Nees]. Ind Crops and Prod 2016 86:1–1.
Agarwal A, Gupta V, Haq SU, Jatav PK, Kothari SL, Kachhwaha S (2019) Assessment of genetic diversity in 29 rose germplasms using SCoT marker. J King Saud Univ Sci 31(4):780–788
Verma KS, ul Haq S, Kachhwaha S, Kothari SL (2017) RAPD and ISSR marker assessment of genetic diversity in Citrullus colocynthis (L.) Schrad: a unique source of germplasm highly adapted to drought and high-temperature stress. 3 Biotech 7(5):288.
Satya P, Karan M, Jana S, Mitra S, Sharma A, Karmakar PG, Ray DP (2015) Start codon targeted (SCoT) polymorphism reveals genetic diversity in wild and domesticated populations of ramie (Boehmeria nivea L. Gaudich.), a premium textile fiber producing species. Meta Gene 3:62–70
Mizusawa L, Kaneko S, Hasegawa M, Isagi Y (2011) Development of nuclear SSRs for the insular shrub Clerodendrum izuinsulare (Verbenaceae) and the widespread C. trichotomum. Am J Bot 98(11):e333–6.
Prasad MP, Sushant S, Chikkaswamy BK (2012) Phytochemical analysis, antioxidant potential, antibacterial activity and molecular characterization of Clerodendrum species. Int J Mol Biol 3(3):71–76
Liu P, Que Y, Pan YB (2011) Highly polymorphic microsatellite DNA markers for sugarcane germplasm evaluation and variety identity testing. Sugar Tech 13(2):129–136
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98(1):107–112
Rohlf FJ (1998) NTSYS: numerical taxonomy and multivariate analysis system version 2.02. Applied Biostatistics Inc., Setauket, NY.
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaudoise Sci Nat 44:223–270
Yeh FC, Yang RC, Boyle T (1999) POPGENE version 1.32: Microsoft Windows–based freeware for population genetic analysis, quick user guide. Center for International Forestry Research, University of Alberta, Edmonton, Alberta, Canada.
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70(12):3321–3323
Pritchard J, Wen X, Falush D (2012) Documentation for structure software: Version 2.3. 4. https://www.pritchbsd.uchicago.edu/structure.html
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Peakall RO, Smouse PE. (2012) GenALEx 6.5 genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28(19):2537–2539.
Vinceti B, Loo J, Gaisberger H, Van Zonneveld MJ, Schueler S, Konrad H (2013) Conservation priorities for Prunus africana defined with the aid of spatial analysis of genetic data and climatic variables. PLoS ONE 8(3):e59987
Pandey MK, Gautami B, Jayakumar T, Sriswathi M, Upadhyaya HD, Gowda MVC, Radhakrishnan T, Bertioli D, Knapp SJ, Cook DR, Varshney RK (2012) Highly informative genic and genomic SSR markers to facilitate molecular breeding in cultivated groundnut (Arachis hypogaea). Plant Breed 131:139–147
Guo DL, Zhang JY, Liu CH (2012) Genetic diversity in some grape varieties revealed by SCoT analyses. Mol Biol Rep 39:5307–5313
Amirmoradi B, Talebi R, Karami E (2012) Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Syst Evol 298:1679–1688
Xiong F, Zhong R, Han Z, Jiang J, He L, Zhuang W, Tang R (2011) Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38:3487–3494
Luo C, He XH, Chen H, Hu Y, Ou SJ (2012) Genetic relationship and diversity of Mangifera indica L.: revealed through SCoT analysis. Genet Resour Crop Evol 59:1505–1515
Muthusamy S, Kanagarajan S, Ponnusamy S (2008) Efficiency of RAPD and ISSR markers system in accessing genetic variation of rice bean (Vigna umbellata) landraces. Electron J Biotechnol 11(3):32–41
Sheeja TE, Sabeesh C, Shabna OV, Shalini RS, Krishnamoorthy B (2013) Genetic diversity analysis of Myristica and related genera using RAPD and ISSR markers. JOSAC 22:38–46
Ying-Xiong QI, Xin-Wen ZH, Cheng-Xin FU, Chan YS (2005) A preliminary study of genetic variation in the endangered, Chinese endemic species Dysosma versipellis (Berberidaceae). Bot Bull Acad Sinica 46.
Rafizadeh A, Koohi-Dehkordi M, Sorkheh K (2018) Molecular insights of genetic variation in milk thistle (Silybum marianum [L.] Gaertn.) populations collected from southwest Iran. Mol Bio Rep 45(4):601–9.
Wright S (1969) Evolution and genetics of populations: The theory of gene frequencies. Chicago Univ. Press, Chicago, USA, Dogan NY, Kurt P, Osma E. Determination of genetic diversity of Sonchuser zincanicus Matthews (Asteraceae), a Critically Endangered plant endemic to Turkey using RAPD markers. Biharean Biologist. 2018 Dec 1;12(2).
Collard BCY, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene targeted markers in plants. Plant Mol Biol Report 27:86–93
Gorji AM, Poczai P, Polgar Z, Taller J (2011) Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am J Potato Res 88(3):226–237
Shahlaei A, Torabi S, Khosroshahli M (2014) Efficacy of SCoT and ISSR markers in assessment of tomato (Lycopersicum esculentum Mill.) genetic diversity. Int J Biol Sci 5:14–22
Hamidi H, Talebi R, Keshavarzi F (2014) Comparative efficiency of functional gene-based markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived polymorphism (CDDP) with ISSR markers for diagnostic fingerprinting in wheat (Triticum aestivum L.). Cereal Res Commun 42(4):558–67.
Igwe DO, Afiukwa CA, Ubi BE, Ogbu KI, Ojuederie OB, Ude GN (2017) Assessment of genetic diversity in Vigna unguiculata L. (Walp) accessions using inter-simple sequence repeat (ISSR) and start codon targeted (SCoT) polymorphic markers. BMC Gent 18(1):98.
Hao Q, Lu C, Zhu Y, Xiao Y, Gu X (2018) Numerical investigation into the evolution of groundwater flow and solute transport in the Eastern Qaidam Basin since the Last Glacial Period. Geofluids 2018.
Costa R, Pereira G, Garrido I, Tavares-de-Sousa MM, Espinosa F (2016) Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify orchard grass (Dactylis glomerata L.) germplasm variations. PloS one 11(4):e0152972.
Ikegami H, Nogata H, Hirashima K, Awamura M, Nakahara T (2009) Analysis of genetic diversity among European and Asian fig varieties (Ficus carica L.) using ISSR, RAPD, and SSR markers. Genet Resour Crop Evol 56:201–209
Satish L, Shilpha J, Pandian S, Rency AS, Rathinapriya P, Ceasar SA, Largia MJ, Kumar AA, Ramesh M (2016) Analysis of genetic variation in sorghum (Sorghum bicolor (L.) Moench) genotypes with various agronomical traits using SPAR methods. Gene 576:581–585
Acknowledgements
Barbi Gogoi is thankful to Dr. G.N. Sastry, Director, CSIR-North-East Institute of Science and Technology, Jorhat, Assam, India and Dr. S.P. Saikia and Dr. S.B. Wann for their consistent support in completing the manuscript.
Funding
This study is financially supported by “CSIR-Direct SRF” fellowship under CSIR-HR Division, New Delhi.
Author information
Authors and Affiliations
Contributions
BG performed the experiment, analysed data and wrote the manuscript. SPS designed the project and wrote the manuscript and SBW provided technical support.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interests.
Research involving human participants or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gogoi, B., Wann, S.B. & Saikia, S.P. Comparative assessment of ISSR, RAPD, and SCoT markers for genetic diversity in Clerodendrum species of North East India. Mol Biol Rep 47, 7365–7377 (2020). https://doi.org/10.1007/s11033-020-05792-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05792-x