Abstract
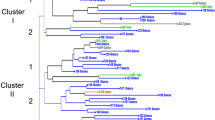
Nineteen fig varieties and lines from Europe and Asia have been fingerprinted by ISSR, RAPD, and SSR markers, respectively, using 13, 19, and 13 primer combinations. All primers produced 258 loci, with the highest number of loci (119) generated by RAPD (R p: 48.42). Clustering analysis was applied to the three marker datasets to elucidate the genetic structure and relationships among these varieties. Mean genetic similarities were 0.787, 0.717, and 0.749, respectively, as determined using ISSR, RAPD, and SSR. Each marker system produced incompletely separated clusters, although a weak binding group based on race type appeared in the combined dataset. Comparisons of coefficients revealed no correlation between different similarity matrices; congruence was observed between similarity matrices and co-phenetic matrices in all markers. Analysis of molecular variance (AMOVA) showed that most of the total polymorphism was attributable to within-group variance (ISSRs + RAPDs, 97.41%; SSRs, 90.18%). These results suggest that the genetic diversity of this fig population is low and that multiple marker utilization is critical to estimate the relatedness of figs at the variety level. Additionally, it was presumed that ‘Houraihi’, the oldest variety in Japan, was disseminated independently of other foreign varieties in the 17th century or before then.



Similar content being viewed by others
References
Aksoy U (1998) Why fig? An old taste and a new perspective. Acta Hortic 480:25–26
Bohn M, Utz HF, Melchinger AE (1999) Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs and SSRs and their use for predicting progeny variance. Crop Sci 39:228–237
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Cabrita LF, Aksoy U, Hepaksoy S, Leitao JM (2001) Suitability of isozyme, RAPD and AFLP markers to assess genetic differences and relatedness among fig (Ficus carica L.) clones. Sci Hortic 87:261–273
Condit IJ (1947) The fig. Chronica Botanica Co., Waltham
De Masi L, Castaldo D, Galano G, Minasi P, Laratta B (2005) Genotyping of fig (Ficus carica L.) via RAPD markers. J Sci Food Agric 85:2235–2242
Gilbert JE, Lewis RV, Wilkinson MJ, Caligari PDS (1999) Developing an appropriate strategy to assess genetic variability in plant germplasm collections. Theor Appl Genet 98:1125–1131
Giraldo E, Viruel MA, Lopez-Corrales M, Hormaza JI (2005) Characterisation and cross-species transferability of microsatellites in the common fig (Ficus carica L.). J Hortic Sci Biotechnol 80:217–224
IBPGR (1986) Genetic resources of tropical and sub-tropical fruits and nuts. IBPGR Rome, pp 63–69
Jones DA (1972) Blood samples: probability of discrimination. J Forensic Sci Soc 12:355–359
Khadari B, Hochu I, Satoni S, Kjellberg F (2001) Identification and characterization of microsatellite loci in the common fig (Ficus carica L.) and representative species of the genus Ficus. Mol Ecol Notes 1:191–193
Khadari B, Ater M, Mamouni A, Roger JP, Kjellberg F (2004) Molecular characterization of Moroccan fig germplasm using inter simple sequence repeat and simple sequence repeat markers to establish a reference collection. HortScience 40(1):29–32
Khadari B, Grout C, Santoni S, Kjellberg F (2005) Contrasted genetic diversity and differentiation among Mediterranean populations of Ficus carica L.: a study using mtDNA RFLP. Genet Resour Crop Evol 52:97–109
Khoshbakht K, Hammer K (2006) Savadkouh (Iran) – an evolutionary center for fruit trees and shrubs. Genet Resour Crop Evol 53:641–651
Lamboy WF, Alpha CG (1998) Using simple sequence repeats (SSRs) for DNA fingerprinting germplasm accessions of grape (Vitis L.) species. J Am Soc Hortic Sci 123:182–188
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M, Li WH (1979) Mathematical model for studying genetical variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 74:5267–5273
Papadopoulou K, Ehaliotis C, Tourna M, Kastanis P, Karydis I, Zervakis G (2002) Genetic relatedness among dioecious Ficus carica L. cultivars by random amplified polymorphic DNA analysis, and evaluation of agronomic and morphological characters. Genetica 114:183–194
Pejic I, Ajmone-Marsan P, Morgante M, Kozumplick V, Castiglioni P, Taramino G et al (1998) Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs and AFLPs. Theor Appl Genet 97:1248–1255
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S et al (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Rohlf FJ (2006) NTSYSpc: numerical taxonomy and multivariate analysis system. Ver. 2.2, Exeter Software, NY
Sadder MT, Ateyyeh AF (2006) Molecular assessment of polymorphism among local Jordanian genotypes of common fig (Ficus carica L.). Sci Hortic Amsterdam 107:347–351
Salhi-Hannachi A, Trifi M, Zehdi S, Hedhi J, Mars M, Rhouma A et al (2004) Inter simple sequence repeat fingerprints to access genetic diversity in Tunisian fig (Ficus carica L.) germplasm. Genet Resour Crop Evol 51:269–275
Salhi-Hannachi A, Chatti K, Mars M, Marrakchi M, Trifi M (2005) Comparative analysis of genetic diversity in two Tunisian collections of fig cultivars based on random amplified polymorphic DNA and inter simple sequence repeats fingerprints. Genet Resour Crop Evol 52:563–573
Sehgal D, Raina SN (2005) Genotyping safflower (Carthamus tinctorius) cultivars by DNA fingerprints. Euphytica 146:67–76.
Storey WB et al (1975) Figs. In: Janick J, Moore J (eds) Advances in fruit breeding. Purdue University Press, Indiana, pp 568–589
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Zukovskij PM (1950) Ficus. Cultivated plants and their wild relatives. State Publishing House Sov Sci Mosc 1950:58–59
Acknowledgement
We are grateful to Dr. Tanisaka for the critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ikegami, H., Nogata, H., Hirashima, K. et al. Analysis of genetic diversity among European and Asian fig varieties (Ficus carica L.) using ISSR, RAPD, and SSR markers. Genet Resour Crop Evol 56, 201–209 (2009). https://doi.org/10.1007/s10722-008-9355-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-008-9355-5