Abstract
Passion fruit (Passiflora edulis), an important tropical and subtropical fruit, has a high edible and medicinal value. Stem rot disease is one of the most important diseases of passion fruit. An effective way for control and prevention of this disease is to identify the genes associated with resistance to this disease. Quantitative real-time PCR (RT-qPCR) has mainly been widely applied to detect gene expression because of its simplicity, fastness, low cost and high sensitivity. One of the requirements for RT-qPCR is the availability of suitable reference genes for normalization of gene expression. However, currently, no Passiflora edulis reference genes have been identified andthus it has hindered the gene expression studies in this plant. The present study aimed to address this issue. We analyzed sixteen candidate reference genes, including nine common (GAPDH, UBQ, ACT1, ACT2, EF-1α-1, EF-1α-2, TUA, NADP, and GBP) and seven novel genes (C13615, C24590, C27182, C10445, C21209, C22199, and C22526), in different tissues (stem, leaf, flower and fruit) of two accessions under stem rot condition. We calculated the expression stability in twenty-four samples using the ΔCt, GeNorm, NormFinder, BestKeeper and RefFinder. The results showed that both C21209 and EF-1α-2 were sufficient to normalize gene expression under stem rot, whereas the commonly used reference genes, GAPDH and UBQ, were the least stable ones. The expression patterns of PeUFC under stem rot condition normalized by stable and unstable reference genes indicated the suitability of using the optimal reference genes. To our knowledge, this is the first systematic study of reference genes in Passiflora edulis, which identified a number of reliable reference genes suitable for gene expression studies in Passiflora edulis by RT-qPCR.






Similar content being viewed by others
References
Munhoz CF, Costa ZP, Cauz-santos LA et al (2018) A gene-rich fraction analysis of the Passiflora edulis genome reveals highly conserved microsyntenic regions with two related Malpighiales species. Sci Rep 8:13024. https://doi.org/10.1038/s41598-018-31330-8
Wong ML, Medrano JF (2005) Real-time PCR for mRNA quantitation. Biotechniques 39:75–85. https://doi.org/10.2144/05391RV01
Kozera B, Rapacz M (2013) Reference genes in real-time PCR. J Appl Genet 54:391–406. https://doi.org/10.1007/s13353-013-0173-x
Rapacz M, Stępień A, Skorupa K (2012) Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiol Plant 34:1723–1733. https://doi.org/10.1007/s11738-012-0967-1
Monteiro F, Sebastiana M, Pais MS et al (2013) Reference gene selection and validation for the early responses to downy mildew infection in susceptible and resistant Vitis vinifera cultivars. PLoS ONE 8:e72998. https://doi.org/10.1371/journal.pone.0072998
Li QF, Sun SM, Yuan DY et al (2010) Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep 28:49–57. https://doi.org/10.1007/s11105-009-0124-1
Zhao Y, Luo J, Xu S et al (2016) Selection of reference genes for gene expression normalization in Peucedanum praeruptorum Dunn under abiotic stresses, hormone treatments and different tissues. PLoS ONE 11:e0152356. https://doi.org/10.1371/journal.pone.0152356
Carvalho KD, Bespalhok FJ, Santos TD et al (2013) Nitrogen starvation, salt and heat stress in coffee (Coffea arabica L.): identification and validation of new genes for qPCR normalization. Mol Biotechnol 53:315–325. https://doi.org/10.1007/s12033-012-9529-4
Wan H, Zhao Z, Qian C et al (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261. https://doi.org/10.1016/j.ab.2009.12.008
Chen H, Yang ZQ, Hu Y et al (2016) Reference genes selection for quantitative gene expression studies in Pinus massoniana L. Trees 30:685–696. https://doi.org/10.1007/s00468-015-1311-3
Liu D, Shi L, Han C et al (2012) Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS ONE 7:e46451. https://doi.org/10.1371/journal.pone.0046451
Xu M, Li A, Teng Y et al (2019) Exploring the adaptive mechanism of Passiflora edulis in karst areas via an integrative analysis of nutrient elements and transcriptional profiles. BMC Plant Biol 19:185. https://doi.org/10.1186/s12870-019-1797-8
Liu S, Li AD, Chen CH et al (2017) De Novo transcriptome sequencing in Passiflora edulis sims to identify genes and signaling pathways involved in cold tolerance. Forests 8:435. https://doi.org/10.3390/f8110435
Araya S, Martins AM, Junqueira N et al (2017) Microsatellite marker development by partial sequencing of the sour passion fruit genome (Passiflora edulis Sims). BMC Genomics 18:549. https://doi.org/10.1186/s12864-017-3881-5
Costa ZD, Munhoz CF, Vieira M (2017) Report on the development of putative functional SSR and SNP markers in passion fruits. BMC Res Notes 10:445. https://doi.org/10.1186/s13104-017-2771-x
Munhoz CF, Santos AA, Arenhart RA, Santini L et al (2015) Analysis of plant gene expression during passion fruit–xanthomonas axonopodis interaction implicates lipoxygenase 2 in host defence. Ann Appl Biol 167:135–155. https://doi.org/10.1111/aab.12215
Xie F, Sun G, Stiller JW et al (2011) Genome-wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLoS ONE 6:e26980. https://doi.org/10.1371/journal.pone.0026980
Vandesompele J, De PK, Pattyn F et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:1–11. https://doi.org/10.1186/gb-2002-3-7-research0034
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496
Pfaffl MW, Tichopad A, Prgomet C et al (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. https://doi.org/10.1023/b:bile.0000019559.84305.47
Xie F, Xiao P, Chen D et al (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80:75–84. https://doi.org/10.1007/s11103-012-9885-2
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12:323. https://doi.org/10.1186/1471-2105-12-323
Ye J, Zhong T, Zhang D et al (2019) The auxin-regulated protein ZmAuxRP1 coordinates the balance between root growth and stalk rot disease resistance in maize. Mol Plant 12:360–373. https://doi.org/10.1016/j.molp.2018.10.005
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3–new capabilities and interfaces. Nucleic Acids Res 40:e115. https://doi.org/10.1093/nar/gks596
Sun HF, Meng YP, Cui GM et al (2009) Selection of housekeeping genes for gene expression studies on the development of fruit bearing shoots in Chinese jujube (Ziziphus jujube Mill.). Mol Biol Rep 36:2183–2190. https://doi.org/10.1007/s11033-008-9433-y
Mughal BB, Leemans M, Spirhanzlova P et al (2018) Reference gene identification and validation for quantitative real-time PCR studies in developing Xenopus laevis. Sci Rep 8:496. https://doi.org/10.1038/s41598-017-18684-1
Karuppaiya P, Yan XX, Liao W et al (2017) Identification and validation of superior reference gene for gene expression normalization via RT-qPCR in staminate and pistillate flowers of Jatropha curcas-A biodiesel plant. PLoS ONE 12:e0172460. https://doi.org/10.1371/journal.pone.0172460
Du W, Hu F, Yuan S et al (2019) Selection of reference genes for quantitative real-time PCR analysis of photosynthesis-related genes expression in Lilium regale. Physiol Mol Biol Plants 25:1497–1506. https://doi.org/10.1007/s12298-019-00707-y
Dos Santos CP, Da Cruz SK, Batista MC et al (2019) Identification and evaluation of reference genes for reliable normalization of real-time quantitative PCR data in acerola fruit, leaf, and flower. Mol Biol Rep 47:953–965. https://doi.org/10.1007/s11033-019-05187-7
Li Z, Lu H, He Z et al (2019) Selection of appropriate reference genes for quantitative real-time reverse transcription PCR in Betula platyphylla under salt and osmotic stress conditions. PLoS ONE 14:e0225926. https://doi.org/10.1371/journal.pone.0225926
Wang X, Wu Z, Bao W et al (2019) Identification and evaluation of reference genes for quantitative real-time PCR analysis in Polygonum cuspidatum based on transcriptome data. BMC Plant Biol 19:498. https://doi.org/10.1186/s12870-019-2108-0
Hu X, Zhang L, Nan S et al (2018) Selection and validation of reference genes for quantitative real-time PCR in Artemisia sphaerocephala based on transcriptome sequence data. Gene 657:39–49. https://doi.org/10.1016/j.gene.2018.03.004
Gao D, Kong F, Sun P et al (2018) Transcriptome-wide identification of optimal reference genes for expression analysis of Pyropia yezoensis responses to abiotic stress. BMC Genomics 19:251. https://doi.org/10.1186/s12864-018-4643-8
Wang G, Tian C, Wang Y et al (2019) Selection of reliable reference genes for quantitative RT-PCR in garlic under salt stress. Peer J 7:e7319. https://doi.org/10.7717/peerj.7319
Feng K, Liu JX, Xing GM et al (2019) Selection of appropriate reference genes for RT-qPCR analysis under abiotic stress and hormone treatment in celery. Peer J 7:e7925. https://doi.org/10.7717/peerj.7925
Cheng T, Zhu F, Sheng J et al (2019) Selection of suitable reference genes for quantitive real-time PCR normalization in Miscanthus lutarioriparia. Mol Biol Rep 46:4545–4553. https://doi.org/10.1007/s11033-019-04910-8
Zhang K, Li M, Cao S et al (2019) Selection and validation of reference genes for target gene analysis with quantitative real-time PCR in the leaves and roots of Carex rigescens under abiotic stress. Ecotoxicol Environ Saf 168:127–137. https://doi.org/10.1016/j.ecoenv.2018.10.049
Phule AS, Barbadikar KM, Madhav MS et al (2018) Genes encoding membrane proteins showed stable expression in rice under aerobic condition: novel set of reference genes for expression studies. 3 Biotech 8:383. https://doi.org/10.1007/s13205-018-1406-9
Borkowska P, Zielińska A, Paul-samojedny M et al (2020) Evaluation of reference genes for quantitative real-time PCR in Wharton's Jelly-derived mesenchymal stem cells after lentiviral transduction and differentiation. Mol Biol Rep 47:1107–1115. https://doi.org/10.1007/s11033-019-05207-6
Li C, Hu L, Wang X et al (2019) Selection of reliable reference genes for gene expression analysis in seeds at different developmental stages and across various tissues in Paeonia ostii. Mol Biol Rep 46:6003–6011. https://doi.org/10.1007/s11033-019-05036-7
Hou F, Li S, Wang J et al (2017) Identification and validation of reference genes for quantitative real-time PCR studies in long yellow daylily Hemerocallis citrina Borani. PLoS ONE 12:e0174933
Wang C, Yang Q, Wang W et al (2017) A transposon-directed epigenetic change in ZmCCT underlies quantitative resistance to Gibberella stalk rot in maize. New Phytol 215:1503–1515. https://doi.org/10.1111/nph.14688
Shen L, Zhong T, Wang L et al (2019) Characterization the role of a UFC homolog, AtAuxRP3, in the regulation of Arabidopsis seedling growth and stress response. J Plant Physiol 240:152990. https://doi.org/10.1016/j.jplph.2019.152990
Huot B, Yao J, Montgomery BL et al (2014) Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant 7:1267–1287. https://doi.org/10.1093/mp/ssu049
Wang W, Wang ZY (2014) At the intersection of plant growth and immunity. Cell Host Microbe 15:400–402. https://doi.org/10.1016/j.chom.2014.03.014
Acknowledgements
This work was supported by Guangxi Natural Science Foundation of China (2018GXNSFBA281024, 2019GXNSFAA245002), Guangxi’s Ministry of Science and Technology (AB18294007), Guangxi Academy of Agricultural Sciences (2018YT19, TS2016010). Thanks to Xu et al. for the transcriptome data.
Author information
Authors and Affiliations
Contributions
YW, HM and XY. contributed to study design, QT contributed to qPCR analysis, WH and JL. contributed to data analysis, XX. contributed to primer design, YW. wrote this manuscript, XY. revised the manuscript. All authors read and approve the paper.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
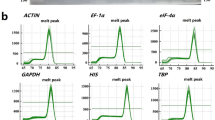
Primer specificity and melting curve analysis of the sixteen candidate reference genes. (a-p): C13615, C24590, C27182, C10445, C21209, C22199, C22526, GAPDH, UBQ, Actin1, Actin2, EF-1α-1, EF-1α-2, TUA, NADP, and GBP. Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, Y., Tian, Q., Huang, W. et al. Identification and evaluation of reference genes for quantitative real-time PCR analysis in Passiflora edulis under stem rot condition. Mol Biol Rep 47, 2951–2962 (2020). https://doi.org/10.1007/s11033-020-05385-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05385-8