Abstract
Miscanthus lutarioriparia, which is found widespread in China, has attracted great attention as a most potential bioenergy plant for years. The quantitative real time PCR (RT-qPCR) has appeared as a sensitive and powerful technique to measure gene expression in living organisms during different development stages. In this study, we evaluated ten candidate genes, including 25S ribosomal RNA gene (25S rRNA), actin1 gene (ACT1), carotenoid-binding protein 20 gene (CBP20), glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH), Ubiquitin gene (UBQ), eukaryotic elongation factor 1-αgene (eEF-1α), α-tubulin gene (α-TUB), β-tubulin gene (β-TUB), eukaryotic translation initiation factor 4α-1 gene (eIF-4α) and NAC domain protein gene(NAC) in a series of 30 M. lutarioriparia samples followed by statistical algorithms geNorm and Normfinder to analyze the gene expression stability. The results indicated that eIF-4αand UBQ were the most stable expressed genes while CBP20 showed as the least stable among all the samples. Based on above research, we recommend that at least two top-ranked reference genes should be employed for expression data normalization. The best genes selected in this study will provide a starting point to select reference genes in the future in other tissues and under other experimental conditions in this energy crop candidate.



Similar content being viewed by others
Abbreviations
- RT-qPCR:
-
Quantitative real-time reverse transcription polymerase chain reaction
- 25S rRNA :
-
25S ribosomal RNA
- ACT1 :
-
Actin 1
- CBP20 :
-
Carotenoid-binding protein 20
- GAPDH :
-
Glyceraldehyde-3-phosphate dehydrogenase
- UBQ :
-
Ubiquitin
- eEF-1α :
-
Elongation factor-1α
- αTUB :
-
α Tubulin
- βTUB :
-
β Tubulin
- eIF-4α :
-
Eukaryotic translation initiation factor 4α
- NAC :
-
NAC domain protein
References
Barling A, Swaminathan K, Mitros T, James BT, Morris J, Ngamboma O, Hall MC, Kirkpatrick J, Alabady M, Spence AK (2013) A detailed gene expression study of the Miscanthus genus reveals changes in the transcriptome associated with the rejuvenation of spring rhizomes. BMC Genom 14(1):864
Kim C, Lee T-H, Guo H, Chung SJ, Paterson AH, Kim D-S, Lee G-J (2014) Sequencing of transcriptomes from two Miscanthus species reveals functional specificity in rhizomes, and clarifies evolutionary relationships. BMC Plant Biol 14(1):134
Liu C, Xiao L, Jiang J, Wang W, Gu F, Song D, Yi Z, Jin Y, Li L (2013) Biomass properties from different Miscanthus species. Food Eng Secur 2(1):12–19
Sang T, Zhu W (2011) China’s bioenergy potential. Gcb Bioenerg 3(2):79–90
Lewandowski I, Clifton-Brown J, Scurlock J, Huisman W (2000) Miscanthus: European experience with a novel energy crop. Biomass Bioenerg 19(4):209–227
Xu Q, Zhu C, Fan Y, Song Z, Xing S, Liu W, Yan J, Sang T (2016) Population transcriptomics uncovers the regulation of gene expression variation in adaptation to changing environment. Sci Rep 6:25536
Mi J, Liu W, Yang W, Yan J, Li J, Sang T (2014) Carbon sequestration by Miscanthus energy crops plantations in a broad range semi-arid marginal land in China. Sci Total Environ 496:373–380
Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30(6):503–512
Sun M, Wang Y, Yang D, Wei C, Gao L, Xia T, Shan Y, Luo Y (2010) Reference genes for real-time fluorescence quantitative PCR in Camellia sinensis. Chin Bull Bot 45(5):579–587
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45–e45
Leong DT, Gupta A, Bai HF, Wan G, Yoong LF, Too H-P, Chew FT, Hutmacher DW (2007) Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials 28(2):203–210
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotech Lett 26(6):509–515
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6(6):609–618
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75(2–3):291–295
Wu Z-J, Tian C, Jiang Q, Li X-H, Zhuang J (2016) Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci Rep 6:19748
Lee PD, Sladek R, Greenwood CM, Hudson TJ (2002) Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res 12(2):292–297
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can Res 64(15):5245–5250
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biol 3(7):research0034
Sheng J, Zheng X, Wang J, Zeng X, Zhou F, Jin S, Hu Z, Diao Y (2017) Transcriptomics and proteomics reveal genetic and biological basis of superior biomass crop Miscanthus. Sci Rep 7(1):13777
Le DT, Aldrich DL, Valliyodan B, Watanabe Y, Van Ha C, Nishiyama R, Guttikonda SK, Quach TN, Gutierrez-Gonzalez JJ, Tran L-SP (2012) Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE 7(9):e46487
Yang M, Zhu L, Xu L, Liu Y (2014) Population structure and association mapping of flower-related traits in lotus (Nelumbo Adans.) accessions. Sci Hortic 175:214–222
Huang L, Yan H, Jiang X, Zhang X, Zhang Y, Huang X, Zhang Y, Miao J, Xu B, Frazier T (2014) Evaluation of candidate reference genes for normalization of quantitative RT-PCR in switchgrass under various abiotic stress conditions. BioEnerg Res 7(4):1201–1211
Chen K, Fessehaie A, Arora R (2012) Selection of reference genes for normalizing gene expression during seed priming and germination using qpcr in Zea mays and Spinacia oleracea. Plant Mol Biol Rep 30(2):478–487. https://doi.org/10.1007/s11105-011-0354-x
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345(2):646–651
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6(1):27
Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8(1):131
Singh R, Green MR (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259(5093):365–368
Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chuang DM (1996) Evidence that glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. J Neurochem 66(3):928–935
Iskandar HM, Simpson RS, Casu RE, Bonnett GD, Maclean DJ, Manners JM (2004) Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol Biol Rep 22(4):325–337
Gimeno J, Eattock N, Van Deynze A, Blumwald E (2014) Selection and validation of reference genes for gene expression analysis in switchgrass (Panicum virgatum) using quantitative real-time RT-PCR. PLoS ONE 9(3):e91474
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9(1):59
D’haene B, Vandesompele J, Hellemans J (2010) Accurate and objective copy number profiling using real-time quantitative PCR. Methods 50(4):262–270
Zhi-wei Z, Chang-sheng D (2006) The Stability comparison of housekeeping genes as internal standards. Lett Biotechnol 5:807–809
Lilly S, Drummond R, Pearson M, MacDiarmid R (2011) Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol Plant Microbe Interact 24(3):294–304
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399(2):257–261
Løvdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387(2):238–242
Nicot N, Hausman J-F, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56(421):2907–2914
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31571740) and the National High-Tech R&D Program (Grant No. 2012AA101801), Natural Science Foundation of Hubei Province (Grant No. 2013CFA103).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2019_4910_MOESM1_ESM.eps
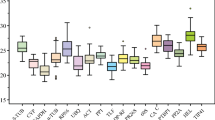
Supplementary material 1 Expression profile of candidate reference genes in different tissues of M. lutarioriparius Ct values in different part of M. lutarioriparius were counted for expression analysis. Red point represent the mean value of the Ct at a certain tissue. Whisskers represent the range of standard errors (EPS 8529 kb)
Rights and permissions
About this article
Cite this article
Cheng, T., Zhu, F., Sheng, J. et al. Selection of suitable reference genes for quantitive real-time PCR normalization in Miscanthus lutarioriparia. Mol Biol Rep 46, 4545–4553 (2019). https://doi.org/10.1007/s11033-019-04910-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04910-8