Abstract
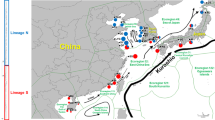
Generally, the gene flow of marine organisms is well maintained, but some local populations of coastal species are genetically differentiated even on a small scale (genetic patchiness). Small-scale isolation can be crucial for understanding genetic diversity within a species. The present study examined the population genetic structure of the sand bubbler crab Scopimera ryukyuensis, which is endemic to the Ryukyu Islands in the northwestern Pacific. A total of 52 haplotypes of mitochondrial cytochrome c oxidase subunit I were recovered from 197 specimens collected from four islands. The haplotype and nucleotide diversities were relatively high in the central Ryukyus (Amami-Oshima and Okinawa Islands) with some exceptions but were low at the southern edge of the geographical distribution of the species, i.e., the southern Ryukyus (Ishigaki and Iriomote Islands). Pairwise FST analysis suggested that the gene flow of S. ryukyuensis was largely restricted. The local populations of the species are differentiated among islands, except for stations on Ishigaki Island and a station on Iriomote Island. Moreover, a clear intra-island population genetic structure was observed within Amami-Oshima and Iriomote Islands, e.g., only 20 km between stations. Small-scale isolation among local populations may be a common tendency for coastal species in the Ryukyu Islands, considering the results of previous studies on corals.



Similar content being viewed by others
References
Cowen RK, Sponaugle S (2009) Larval dispersal and marine population connectivity. Annu Rev Mar Sci 1:443–466. https://doi.org/10.1146/annurev.marine.010908.163757
Nakajima Y, Nishikawa A, Isomura N et al (2009) Genetic connectivity in the broadcast-spawning coral Acropora digitifera analyzed by microsatellite markers on the Sekisei Reef, southwestern Japan. Zool Sci 26:209–215. https://doi.org/10.2108/zsj.26.209
Nakajima Y, Nishikawa A, Iguchi A, Sakai K (2010) Gene flow and genetic diversity of a broadcast-spawning coral in northern peripheral populations. PLoS ONE.https://doi.org/10.1371/journal.pone.0011149
Palumbi SR, Vollmer S, Romano S et al (2012) The role of genes in understanding the evolutionary ecology of reef building corals. Evol Ecol 26:317–335. https://doi.org/10.1007/s10682-011-9517-3
Shinzato C, Mungpakdee S, Arakaki N, Satoh N (2015) Genome-wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Sci Rep 5:18211. https://doi.org/10.1038/srep18211
Zayasu Y, Nakajima Y, Sakai K et al (2016) Unexpectedly complex gradation of coral population structure in the Nansei Islands, Japan. Ecol Evol 6:5491–5505. https://doi.org/10.1002/ece3.2296
Miura O, Kanaya G, Nakai S et al (2017) Ecological and genetic impact of the 2011 Tohoku Earthquake Tsunami on intertidal mud snails. Sci Rep 7:44375. https://doi.org/10.1038/srep44375
Maas DL, Prost S, Bi K et al (2018) Rapid divergence of mussel populations despite incomplete barriers to dispersal. Mol Ecol 27:1556–1571. https://doi.org/10.1111/mec.14556
Wilson RE, Sage GK, Wedemeyer K et al (2019) Micro-geographic population genetic structure within Arctic cod (Boreogadus saida) in Beaufort Sea of Alaska. ICES J Mar Sci 76:1713–1721. https://doi.org/10.1093/icesjms/fsz041
Larson RJ, Julian RM (1999) Spatial and temporal genetic patchiness in marine populations and their implications for fisheries management. Calif Coop Ocean Fish Investig Rep 40:94–99
Kizaki K (1986) Geology and tectonics of the Ryukyu Islands. 125:193–207. https://doi.org/10.2208/proer.33.55
Ota H (1998) Geographic patterns of endemism and speciation in amphibians and reptiles of the Ryukyu Archipelago, Japan, with special reference to their paleogeographical implications. Res Popul Ecol 40:189–204. https://doi.org/10.1007/BF02763404
Yasuma S (2001) Ryukyu Archipelago: biodiversity and generation history of Archipelago. Tokai Library, Tokyo (in Japanese)
Matsumoto T, Kimura M, Nakamura A, Aoki M (1996) Detailed bathymetric features of Tokara and Kerama Gaps in the Ryukyu Arc. J Geogr 105:286–296. https://doi.org/10.5026/jgeography.105.3_286 (in Japanese with English abstract)
Qiu B (2001) Kuroshio and Oyashio Currents. In: Steele J, Thorpe S, Turekian K (eds) Encyclopedia of ocean sciences. Academic, Cambridge, pp 1413–1425
Qiu B, Imasato N (1990) A numerical study on the formation of the Kuroshio Counter Current and the Kuroshio Branch Current in the East China Sea. Cont Shelf Res 10:165–184. https://doi.org/10.1016/0278-4343(90)90028-K
Zhu X, Han I, Park J et al (2003) The Northeastward current southeast of Okinawa Island observed during November 2000 to August 2001. Geophys Res Lett 30:1071. https://doi.org/10.1029/2002GL015867
Ichikawa H, Nakamura H, Nshina A, Higashi M (2004) Variability of northeastward current southeast of northern Ryukyu Islands. J Oceanogr 60:351–363. https://doi.org/10.1023/B:JOCE.0000038341.27622.73
Uchiyama Y, Odani S, Kashima M et al (2018) Influences of the Kuroshio on interisland remote connectivity of corals across the Nansei Archipelago in the East China Sea. J Geophys Res Ocean 123:9245–9265. https://doi.org/10.1029/2018JC014017
Iida M, Zenimoto K, Watanabe S, Kimura S, Tsukamoto K (2010) Larval transport of the amphidromous goby Sicyopterus japonicus by the Kuroshio Current. Coast Mar Sci 34:42–46
Adjeroud M, Tsuchiya M (1999) Genetic variation and clonal structure in the scleractinian coral Pocillopora damicornis in the Ryukyu Archipelago, southern Japan. Mar Biol 134:753–760. https://doi.org/10.1007/s002270050592
Yasuda N, Taquet C, Nagai S et al (2015) Genetic connectivity of the coral-eating sea star Acanthaster planci during the severe outbreak of 2006–2009 in the Society Islands, French Polynesia. Mar Ecol 36:668–678. https://doi.org/10.1111/maec.12175
Zayasu Y, Satoh N, Shinzato C (2018) Genetic diversity of farmed and wild populations of the reef-building coral, Acropora tenuis. Restor Ecol 26:1195–1202. https://doi.org/10.1111/rec.12687
Kojima S, Kamimura S, Kimura T et al (2003) Phylogenetic relationships between the tideland snails Batillaria flectosiphonata in the Ryukyu Islands and B. multiformis in the Japanese Islands. Zool Sci 20:1423–1433. https://doi.org/10.2108/zsj.20.1423
Kojima S, Kamimura S, Iijima A et al (2006) Molecular phylogeny and population structure of tideland snails in the genus Cerithidea around Japan. Mar Biol 149:525–535. https://doi.org/10.1007/s00227-005-0183-2
Yamasaki I, Yoshizaki G, Yokota M et al (2006) Mitochondrial DNA variation and population structure of the Japanese mitten crab Eriocheir japonica. Fish Sci 72:299–309. https://doi.org/10.1111/j.1444-2906.2006.01151.x
Aoki M, Wada K (2013) Genetic structure of the wide-ranging fiddler crab Uca crassipes in the west Pacific region. J Mar Biol Assoc UK 93:789–795. https://doi.org/10.1017/S0025315412001178
Kawane M, Wada K (2015) Genetic population structure of the rare brackish-water crab Ptychognathus ishii Sakai, 1939 (Varunidae) on the Japanese coast. Jpn J Benthol.https://doi.org/10.5179/benthos.70.13
Kawaida S, Nanjo K, Kanai T, Kohno H (2017) Microhabitat differences in crab assemblage structures in a subtropical mangrove estuary on Iriomote Island, southern Japan. Fish Sci 83:1007–1017. https://doi.org/10.1007/s12562-017-1139-4
Wong KJH, Chan BKK, Shih HT (2010) Taxonomy of the sand bubbler crabs Scopimera globosa De Haan, 1835, and S. tuberculata Stimpson, 1858 (Crustacea: Decapoda: Dotillidae) in East Asia, with description of a new species from the Ryukyus, Japan. Zootaxa 2345:43–59. https://doi.org/10.11646/zootaxa.2345.1.4
Irie M, Kawachi A, Ishigami T, Ishikawa T (2005) Study on habitat and territorial competition of crabs which ranges on Amparu Tidal Lagoon. Environ Syst Res 33:55–62. https://doi.org/10.2208/proer.33.55 (in Japanese with English abstract)
Fielder DR (1970) The feeding behaviour of the sand crab Scopimera inflata (Decapoda, Ocypodidae). J Zool Lond 160:35–49. https://doi.org/10.1111/j.1469-7998.1970.tb02896.x
Yutani K, Komori M, Asaeda T (2013) Bioturbation on tidal flat by the burrowing of Ilyoplax pusilla and Scopimera globosa. J Jpn Soc Civ Eng B1 69:1429–1434. https://doi.org/10.2208/jscejhe.69.I_1429 (in Japanese with English Abstract)
Oshima K (1966) Bottom deposits and benthic fauna in Usu Bay, Hokkaido. J Geol Soc Jpn 72:439–449. https://doi.org/10.5575/geosoc.72.439 (in Japanese with English abstract)
Wessel P, Smith WHF, Scharroo R et al (2013) Generic Mapping Tools: improved version released. EOS Trans AGU 94:409–410. https://doi.org/10.1002/2013EO450001
Folmer O, Black M, Hoeh W et al (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299. https://doi.org/10.1371/journal.pone.0013102
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Storey JD, Bass AJ (2019) Bioconductor’s q value package Version 2.18.0. https://www.bioconductor.org/packages/release/bioc/vignettes/qvalue/inst/doc/qvalue.pdf. Accessed 28 Dec 2019
Storey JD (2002) A direct approach to false discovery rates. J R Stat Soc B 64:479–498. https://doi.org/10.1111/1467-9868.00346
R Core Team (2017) R: a language and environment for statistical computing. https://www.R-project.org/. Accessed 27 Jan 2020
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC et al (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Clement M, Snell Q, Walker P et al (2002) Estimating gene genealogies. Parallel Distrib Process Symp Int Proc 2:184. https://doi.org/10.1046/j.1365-294x.2000.01020.x
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210X.12410
Aoki M, Naruse T, Cheng JH et al (2008) Low genetic variability in an endangered population of fiddler crab Uca arcuata on Okinawajima Island: analysis of mitochondrial DNA. Fish Sci 74:330–340. https://doi.org/10.1111/j.1444-2906.2008.01529.x
Lesica P, Allendorf FW (1995) When are peripheral populations valuable for conservation? Conserv Biol 9:753–760. https://doi.org/10.1046/j.1523-1739.1995.09040753.x
Kojima S, Kamimura S, Iijima A et al (2005) Phylogeography of the endangered tideland snail Batillaria zonalis in the Japanese and Ryukyu Islands. Ecol Res 20:686–694. https://doi.org/10.1007/s11284-005-0082-5
Hirase S, Kanno M, Ikeda M, Kijima A (2012) Evidence of the restricted gene flow within a small spatial scale in the Japanese common intertidal goby Chaenogobius annularis. Mar Ecol 33:481–489. https://doi.org/10.1111/j.1439-0485.2012.00512.x
Yamada A, Furukawa F, Wada K (2009) Geographical variations in waving display and barricade-building behaviour, and genetic population structure in the intertidal brachyuran crab (de Haan, 1835). J Nat Hist 43(1–2):17–34
Yatsuzuka K (1957) Study of brachyuran zoea (artificial rearing and development). In: Suehiro Y, Oshima Y, Hiyama Y (eds) Suisangaku Shusei. University of Tokyo Press, Tokyo, pp 571–590 (in Japanese)
Acknowledgements
I am extremely grateful to Hirokazu Abe, Ryutaro Goto, and Masatoshi Matsumasa for providing information about sampling stations on Amami-Oshima Island. I am especially thankful to Hiroaki Fukumori, Hajime Itoh, Shigeaki Kojima, Naoya Ohtsuchi, and Keiji Wada for their insightful suggestions on my research. The anonymous reviewer is thanked for his/her invaluable comments on the earlier version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kobayashi, G. Small-scale population genetic structure of the sand bubbler crab Scopimera ryukyuensis in the Ryukyu Islands, Japan. Mol Biol Rep 47, 2619–2626 (2020). https://doi.org/10.1007/s11033-020-05350-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05350-5