Abstract
Polymorphic variants at the interleukin-2 (IL2) locus affect the risk of several autoimmune disorders. Our aim was to evaluate the association of the four IL2 polymorphisms (rs6822844, rs6534349, rs2069762 and rs3136534) with type 1 diabetes (T1D) in the Polish population, and to correlate them with the serum interleukin-2 levels. 543 unrelated T1D patients and 706 healthy control subjects were enrolled. The minor T allele at rs6822844 was significantly less frequent in T1D compared to controls (p = 0.002; OR 0.71; 95 % CI 0.571–0.880). Likewise, the frequency of the TT genotype was decreased among the affected individuals (p = 0.007). In healthy subjects, stratification according to the rs6822844 genotype revealed significant differences in circulating interleukin-2 (p = 0.037) with the highest levels in TT protective genotypes. Three other IL2 polymorphisms did not display significant differences in allele and genotype distribution. In conclusion, the rs6822844 variant is associated with T1D and may play a functional role, or reflect the influence of another causative genetic variant in linkage disequilibrium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Type 1 diabetes (T1D) results from selective destruction of the insulin-secreting pancreatic beta cells by the autoreactive T lymphocytes. The disease onset is probably triggered by unknown environmental factors (diet, infection) superimposed on specific genetic background in susceptible individuals. To date several genetic loci have been identified which may affect the immune function and thereby contribute to T1D development. The strongest effect was demonstrated for the HLA region (chromosome 6p21), but other genes involved in the immune response also play a role [1]. The IL2 gene, which encodes interleukin-2, a cytokine produced predominately by the activated CD4+ and CD8+ T cells in response to T cell receptor (TCR) stimulation, was one of the strong functional candidates. Plausible association of the IL2 gene with diabetes was first suggested in non-obese diabetic (NOD) mouse model, based on congenic mapping of the Idd3 susceptibility locus [2]. Functional data revealed that IL2 variants in NOD mice might affect the interleukin-2 production by antigen-specific T cells and predispose to organ-specific autoimmunity [3]. These analyses were followed by the first genome-wide association scan in human T1D, which showed a moderate evidence of the association with the IL2 region (4q27) [4]. Rs6534347, an intronic variant of the nearby KIAA1109 gene, was identified by imputation and rs17388568, localized to an intron in the adjacent TENR (ADAD1) gene, revealed the strongest signal based on multilocus analysis [4]. Further sequencing and genotyping of the IL2 region in T1D cohorts revealed the associations with rs3136534, rs6836189 and replicated an association with rs17388568, however, due to extensive linkage disequilibrium (LD) across this locus, the genuine causative variant could not be determined [5].
Early on, interleukin-2 was mainly regarded as a core cytokine in protective cell-mediated immunity, responsible for the clonal expansion of the effector T cells. Nowadays its major and non-redundant action is attributed to the development and homeostasis of the regulatory CD4+CD25+Foxp3+ T (Treg) cells, which ensure the elimination of the self-reactive T lymphocytes [6]. Interleukin-2-deficient mice develop a lethal lymphoproliferative syndrome with massive inflammatory lesions and detectable autoantibodies [7]. There is increasing body of in vitro evidence to support the crucial role of the IL2 signaling pathway in Treg cell function and maintenance of self-tolerance [8]. Therefore, polymorphic variants at the 4q27 locus, which might alter the IL2 expression, could consequently affect the work of the immune system. Indeed, single nucleotide polymorphisms (SNPs) in this region were found to be associated with systemic and organ-specific autoimmune disorders, such as rheumatoid and juvenile idiopathic arthritis, multiple sclerosis, psoriasis, celiac disease, inflammatory bowel disease [9–15]. Allergic conditions—IgE-mediated allergy, asthma and atopic dermatitis—also displayed an association with the IL2 gene variants [16]. Moreover, IL2 SNPs were linked with other immune phenomena, such as measles vaccine-induced immunity and the risk of graft-versus-host disease in patients undergoing hematopoietic stem cell transplantation [17, 18]. However, despite the unequivocal association with other autoimmune disorders and promising GWAS results, 4q27 locus has been rarely examined in the replication studies in different T1D populations to date [9, 10, 19].
The aim of this study was to investigate the association of selected polymorphic variants of the IL2 gene (rs6822844 G/T, rs6534349 A/G, rs2069762 G/T, and rs3136534 A/C) with T1D in the Polish population. We also made an attempt to correlate the genotyping results with serum levels of the interleukin-2, which may represent an intermediate phenotype, directly affected by the genetic factors.
Materials and methods
Selection of investigated IL2 SNPs
The choice of investigated polymorphisms was based upon previous reports showing their involvement in T1D and other autoimmune disorders. Rs6822844, located between the IL2 and the neighboring IL21 gene, was found to be associated with celiac disease, inflammatory bowel disease, psoriasis, juvenile idiopathic and rheumatoid arthritis [9–13, 15]. Rs2060762, located upstream to the IL2 promoter (at −330 site), was previously shown to associate with autoimmune (multiple sclerosis, MS) and allergic conditions [14, 16]. The choice of rs3136534 was supported by the data from the first genome-wide association study in T1D [5]. Finally, rs6534349 was selected as a tagger (r 2 > 0.9) for two other IL2 SNPs, rs2069777 and rs2069779, located in intron 2 and 3, respectively.
Genotyping
The studied cohort consisted of 543 unrelated T1D patients (284 females, 259 males) and 706 healthy control subjects (378 females, 328 males), from the local Polish population of Caucasian origin. Patients were recruited at the Department of Pediatric Diabetes and Obesity. Their mean age (±SD) at disease onset was 8.6 ± 4.5 years. The diagnosis of T1D was based upon World Health Organization criteria and all patients required lifetime insulin substitution. Control samples for the genotyping were obtained from healthy blood donors with negative history of autoimmunity and no signs of autoimmune disorders, recruited at the Regional Blood Transfusion Centre. Their mean age was 38.1 ± 10.2 years.
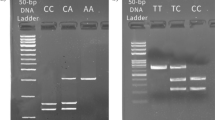
Genomic DNA was extracted from the peripheral blood using Gentra Puregene Blood Kit (Qiagen). Genotyping was performed using the polymerase chain reaction followed by restriction fragment length polymorphism analysis (PCR–RFLP) for SNPs rs6534349, rs2069762 and rs3136534. All fragments were amplified in 30-μl PCR reactions using primers designed with the help of Primer3 online tool, and manufactured by Oligo (Warsaw, Poland) [20]. Primer sequences, PCR conditions, restriction enzymes and fragments are presented in Table 1. Genotyping of the rs6822844 was performed by allelic discrimination analysis using the 7900HT Real-Time PCR System and Taqman assay (C_28983601_10), under the conditions recommended by the manufacturer (Applied Biosystems, Foster City, CA). All genotypes were confirmed in 5 % of samples by direct DNA sequencing with a BigDye Terminator Cycle Sequencing Ready Reaction Kit (ABI Prism 3730 Genetic Analyzer, Foster City, CA). The samples of confirmed genotypes were run as controls in all reactions and 10–12 % of samples were genotyped twice.
Serum interleukin-2 levels
Serum levels of interleukin-2 were determined in a subgroup of 93 consecutive patients with newly diagnosed T1D (mean age 8.9 ± 4.7 years) and 84 age-matched healthy individuals (mean age 8.4 ± 5.3). Samples were collected on the 20–24th day after T1D diagnosis, when the affected subjects reached metabolic homeostasis on insulin substitution, and no signs of inflammation were detectable (confirmed by a negative serum C-reactive protein). Control samples for serum interleukin comparisons were collected from healthy children undergoing routine medical checkup before prophylactic vaccinations. Sera were obtained after overnight fast and stored at −70 °C until analyzed. The cytokine levels were evaluated by means of an ELISA assay (Quantikine® Human IL-2 Immunoassay, R&D Systems, Inc. Minneapolis, MN). The detection limit for interleukin-2 is less than 7 pg/ml and intra- and inter-assay precision CV 2.0–4.3 and 3.7–5.0 %, respectively.
The study was approved by the Ethics Committee at Poznan University of Medical Sciences, and informed consent was obtained from all participants or from their parents in case of minors.
Statistical analysis
Hardy–Weinberg equilibrium was tested for each studied SNP using online calculator available at the Helmholtz Center Munich website (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). Patients and the control group were compared by \({{\upchi}}^{2}\) tests using 2 × 2 and 3 × 2 contingency tables of genotype and allele counts, respectively. The differences were considered significant with a p value <0.05. A Bonferroni correction for multiple test (four independent SNPs) would require a significance threshold of p < 0.0125. Logistic regression was used to determine the allelic odds ratios (ORs) and their 95 % confidence intervals (95 % CI) in case–control comparisons. The calculations were performed using SPSS 18.0 software (SPSS Inc., Chicago, IL). Linkage disequilibrium (LD) measures (r 2) between investigated IL2 markers were assessed with Haploview v.4.1 [21]. The power of the study to detect an association was calculated with the PS Power and Sample Size calculator v.2.1.30 (http://www.mc.vanderbilt.edu/prevmed/ps). Assuming an OR of 0.64, the power of the present study to detect an effect in the T1D cohort was 98 % for rs6822844 [9]. The power to reproduce an OR of 1.54 previously reported for rs2069762 in MS was 92 % [22], and to replicate an OR of 1.11 found for rs3136534 in T1D–only 31 % [5, 9].
Serum levels of interleukin-2 are presented as mean ± standard deviation (±SD) in case of a normal data distribution or as median with their range when the distribution was not normal. Kolmogorov–Smirnov test was used to check for the data normality. Normally distributed interleukin-2 levels in patients and controls were compared using unpaired Student’s t test, while genotype-stratified subgroups displayed non-normal data distribution and hence were analysed by Kruskal–Wallis test. Two-tailed p values < 0.05 were considered statistically significant.
Results
Genotyping
No SNP displayed a significant departure from Hardy–Weinberg equilibrium in the patient and control cohorts (p ≥ 0.074).
The minor T allele at the rs6822844 locus was found in 14.2 % (154 of 1,085) T1D alleles compared to its 18.9 % prevalence (267 of 1,412 alleles) in healthy controls (p = 0.002), yielding a protective OR of 0.71 (95 % CI 0.571–0.880). In line, the frequency of the TT genotype was significantly lower among affected subjects vs. controls (p = 0.007) (Table 2). Both associations survived after correction for multiple testing (4 tests, corrected p-values 0.008 and 0.028, respectively). The other three SNPs: rs6534349, rs2069762, and rs3136534, did not present significant associations in these investigated groups (complete allele and genotype data displayed in Table 2). Linkage disequilibrium measurements revealed very low pairwise r 2 values between rs6822844 and other IL2 SNPs, ranging from 0.01 (rs6822844–rs6534349) to 0.09 (rs6822844–rs3136534). Similar LD results were derived from the HapMap CEU genotype data. Therefore, the haplotype analysis was not performed, as the study of the inferred haplotypes would not add any further information.
Serum interleukin-2 levels
Serum interleukin-2 levels in T1D patients were lower compared to controls however, the difference did not reach statistical significance (8.80 ± 4.95 vs. 10.66 ± 5.42 pg/ml, p = 0.066). Stratification of the control samples according to the rs6822844 genotype revealed significant difference in the interleukin-2 concentration: median 7.94 (range 0.07–17.52) pg/ml for GG, 9.28 (5.25–18.66) pg/ml for GT and 13.80 (6.82–20.83) pg/ml for TT (p = 0.037). On the contrary, circulating interleukin-2 levels in T1D patients were similar in carriers of different rs6822844 genotypes (p = 0.358) (Table 3).
Discussion
The current study indicates an association between rs6822844 polymorphism and T1D in the Polish population. In our analysis, the minor T allele appeared protective, with only 2.9 % of T1D patients carrying the TT genotype compared to 4.1 % of healthy controls. These results confirm formerly published data from the Dutch and Columbian T1D cohorts [9, 10]. On the contrary, in a study of genetic variants shared between T1D and celiac disease, rs6822844 did not reach the threshold of statistical significance in a large British T1D cohort [23]. Likewise, a recent analysis among Spanish T1D subjects revealed only a trend for significance at this SNP [19]. These discrepancies may arise from the population differences, or limited sample size, which might be insufficient to detect a modest effect. However, the direction of this association, with a protective T allele and a major G allele being a risk factor, is common to other autoimmune phenotypes, comprising celiac disease, ulcerative colitis, juvenile idiopathic and rheumatoid arthritis [9, 11–13]. Previous and current findings provide ample evidence for the 4q27 region as a general autoimmunity locus. This 480 kb region harbors four genes: KIAA1109–TENR–IL2–IL21. KIAA1109 is highly expressed in ovaries and brain, TENR expression is testis-specific, whereas IL2 and IL21 encode interleukins, which are both functional candidates in autoimmune diabetes. Interleukin-21 is another T cell-derived cytokine, which enhances the proliferation and effector role of CD8+ T lymphocytes, and is required for generating an inflammatory Th17 response [24]. Priming with interleukin-21 enables autoreactive CD8+ T cells to respond to weak antigens and may induce the diabetes onset in susceptible NOD mice [25].
Rs6822844 is a noncoding SNP, located downstream to the IL2 and approximately 24 kb 5′ of the IL21 gene. Based on sequence homology, it was suggested that the rs6822844-carrying fragment might encode a micro-RNA precursor [10]. A substitution of the conserved G allele by the minor T variant could hamper miRNA synthesis, and consequently influence the expression of other genes. In fact, the true role of this SNP remains to be characterized. One may not exclude that a genuine causal variant, which affects the immune function and contributes to autoimmunity, is located elsewhere within a cluster of several SNPs in the 4q27 region. Rs6822844 was reported as a perfect proxy for the five-SNP haplotype (comprising rs4505484, rs11732095, rs6822844, rs4492018 and rs1398553) that is most associated with autoimmune disease [9]. In contrast, LD between rs6822844 and another T1D-associated SNP, rs17388568, is low (r 2 = 0.09) hence both variants might exert an independent effect. However, localization of rs17388568 in intron 8 of the TENR gene makes its direct influence on immune system rather unlikely.
We did not find any association with three other studied SNPs. Our analysis was underpowered to reliably detect an effect of rs3136534. In contrast, the power of this study was adequate to investigate rs2069762, a polymorphic variant consistently associated with MS [22]. Although T1D and MS are both autoimmune conditions, disturbances of different immune pathways might prevail in their development. For instance, distinct causal SNPs in the 4q27 locus could affect the expression of IL2 and IL21. On the other hand, if susceptibility variants located within the same locus do not overlap, this may reflect the causative role of yet another SNP, which remains in LD. To elucidate this matter, the region should be thoroughly resequenced and further genotyped in large cohorts in order to identify most plausible SNPs for functional analyses.
Interleukin-2 plays a key role in thymic differentiation and peripheral expansion of CD4 + CD25 + Tregs. Polymorphic variants of the genes encoding interleukin-2 and its receptor subunits might enhance/impair their expression and therefore affect the activity of the IL2 signaling pathway. This in turn could influence the function of the CD4+CD25+ Tregs and eventually, promote the development of autoimmune disorders. In fact, SNPs located within the extended murine IL2 promoter, which associate with autoimmune disease, were found to alter the gene transcription in CD4+ T cells [26] and diminished interleukin-2 synthesis was reported for a susceptible haplotype in NOD mice [3]. In humans, the presence of an autoimmunity-associated IL2RA haplotype correlates with reduced interleukin-2 responsiveness in stimulated CD4+ T cells and decreased ability of Tregs to suppress the proliferation of autologous effector T cells [8].
In our study we limited the analysis to serum levels of interleukin-2 in patients and controls, and thereafter we stratified the results according to IL2 genotype. Interleukin-2 is a cardinal regulatory cytokine in the immune system, but its pleiotropic role may impair reliable distinction of the genuine effects from secondary changes related with the disease. This is the most likely reason of major inconsistencies between former studies, which revealed various interleukin-2 levels among T1D subjects compared to healthy controls [27–29]. A trend for lower circulating interleukin-2 in T1D was found in our study. This observation remains in line with most previous analyses and is also corroborated by the results of the peripheral blood mononuclear cell ex vivo cultures [27, 28, 30–32]. However, the experiments, which investigated in vitro synthesis of the interleukin-2 in a longitudinal way, indicate that its peak was similar but considerably delayed in T1D subjects compared to controls [33]. These observations support a hypothesis of impaired interleukin-2 secretory pattern among patients with T1D. Of note, the interleukin-2 production by the T1D lymphocytes appears independent of the degree of the metabolic control, and is probably regulated at a pretranslational stage [30, 31]. Moreover, decreased serum interleukin-2 concentrations were also reported in diabetic patients with long-lasting disease, when the active autoimmune reaction has been attenuated [28]. Therefore, at least partial modulation of the interleukin-2 synthesis could be attributed to genetic factors [32]. Former analyses revealed an association between serum cytokine levels and a polymorphic variant upstream to the IL2 gene promoter (−330 T/G, rs2060762) assessed in MS and in neuroendocrine tumors [14, 34]. Although its position makes rs2060762 a plausible functional candidate, we were not able to confirm this correlation in our cohort (data not shown, p > 0.05). Instead, significant difference in circulating interleukin-2 was found among healthy carriers of three rs6822844 genotypes, and the lowest cytokine levels were present in subjects with the T1D risk GG genotype (Table 3). However, the statistical significance of this finding is borderline. Additionally, this relation was no more significant when evaluated among T1D subjects (p = 0.358), possibly due to overlying immune phenomena connected with the recent disease onset. Replication studies in larger cohorts, as well as experimental investigation of the IL2 gene expression at the mRNA level would be mandatory to confirm these findings. Considering the location of the rs6822844 with regard to the IL2 gene, this SNP seems rather unlikely regulator of the gene expression, however, another unidentified polymorphism in LD may directly affect IL2 regulation. Gene expression usually undergoes complex, multifactorial influence, but similar genotype-intermediate phenotype relationship was reported for the IL2RA gene, which encodes high-affinity alpha subunit of the interleukin-2 receptor (CD25 molecule). Its polymorphic variants are strongly associated with T1D and affect the soluble receptor levels, although discrepancies in the effect of the IL2RA genotype on serum measurements were detected in different autoimmune conditions [35].
In conclusion, the present study confirms the association of the rs6822844 with T1D in another Caucasian population. Furthermore, genotype-related differences in serum interleukin-2 levels suggest plausible functional role of this polymorphism. However, other nearby SNPs, which remain in LD with rs6822844, may play the true causative role. Further functional studies are essential to dissect downstream effects of the molecular variants at this locus and the mechanisms of their contribution to increased risk for T1D and other autoimmune pathology.
References
Pociot F, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G et al (2010) Genetics of type 1 diabetes: what’s next? Diabetes 59:1561–1571
Denny P, Lord CJ, Hill NJ, Goy JV, Levy ER, Podolin PL et al (1997) Mapping of the IDDM locus Idd3 to a 0.35cM interval containing the interleukin-2 gene. Diabetes 46:695–700
Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE et al (2007) Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet 39:329–337
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678
Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V et al (2007) Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Net Genet 39:857–864
Malek TR, Castro I (2010) Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33:153–165
Sadlack B, Lohler J, Schorle H, Klebb G, Haber H, Sickel E et al (1995) Generalized autoimmune disease in interleukin-2-deficient mice is triggered by an uncontrolled activation and proliferation of CD4+ T cells. Eur J Immunol 25:3053–3059
Garg G, Tyler JR, Yang JH, Citler AJ, Downes K, Pekalski M et al (2012) Type 1 diabetes-associated IL2RA variation lowers IL-2 signaling and contributes to diminished CD4+CD25+regulatory T cell function. J Immunol 188:4644–4653
Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B et al (2007) Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet 81:1284–1288
Maiti AK, Kim-Howard X, Viswanathan P, Guillen L, Rojas-Villarraga A, Deshmukh H et al (2010) Confirmation of an association between rs6822844 at the IL2-IL21 region and multiple autoimmune diseases: evidence of a general susceptibility locus. Arthr Rheum 62:323–329
Van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M et al (2007) A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Net Genet 39:827–829
Albers HM, Kurreeman FA, Stoeken-Rijsbergen G, Brinkman DM, Kamphuis SS, van Rossum MA et al (2009) Association of the autoimmunity locus 4q27 with juvenile idiopathic arthritis. Arthr Rheum 60:901–904
Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J et al (2009) Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut 58:779–804
Matesanz F, Fedetz M, Leyva L, Delgardo C, Fernandez O, Alcina A (2004) Effects of the multiple sclerosis-associated -330 promoter polymorphism in IL2 allelic expression. J Neuroimmunol 148:212–217
Warren RB, Smith RL, Flynn E, Bowes J, UKRAG Consortium, Eyre S et al (2011) A systematic investigation of confirmed autoimmune loci in early-onset psoriasis reveals an association with IL2/IL21. Br J Dermatol 164:660–664
Christensen U, Haagerup A, Binderup HG, Vestbo J, Kruse TA, Borglum AD (2006) Family based association analysis of the IL2 and IL15 genes in allergic disorders. Eur J Hum Genet 14:227–235
Haralambieva IH, Ovsyannikova IG, Kennedy RB, Vierkant RA, Pankratz VS, Jacobson RM et al (2011) Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine 29:7883–7895
Harkensee C, Oka A, Onizuka M, Middleton PG, Inoko H, Hirayasy K et al (2012) Single nucleotide polymorphisms and outcome risk in unrelated mismatched hematopoietic stem cell transplantation: an extrapolation study. Blood 199:6365–6372
Espino-Paisan L, De La Calle H, Fernandez-Arquero M, Figueredo MA, De La Concha EG, Urcelay E et al (2011) Study of polymorphisms in 4q27, 10p15 and 22q13 regions in autoantibodies stratified type 1 diabetes. Autoimmunity 44:624–630
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualisation of LD and haplotype maps. Bioinformatics 21:263–265
Matesanz F, Fedetz M, Collado-Romero M, Fernandez O, Guerrero M, Delgardo C et al (2001) Allelic expression and interleukin-2 polymorphisms in multiple sclerosis. J Neuroimmunol 119:101–105
Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH et al (2008) Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med 359:2767–2777
Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L et al (2007) Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448:480–483
Ramanathan S, Dubois S, Chen XL, Leblanc C, Ohashi PS, Ilangumaran S (2011) Exposure to IL-15 and IL-21 enables autoreactive CD8 T cells to respond to weak antigens and cause disease in a mouse model of autoimmune diabetes. J Immunol 186:5131–5141
Del Rio R, Noubade R, Subramanian M, Saligrama N, Diehl S, Rincon M et al (2008) SNPs upstream of the minimal promoter control IL-2 expression and are candidates for the immune disease-susceptibility locus Aod2/Idd3/Eae3. Genes Immun 9:115–121
Kaye WA, Adri MN, Soeldner JS, Rabinowe SL, Kaldany A, Kahn CR et al (1986) Acquired defect in interleukin-2 production in patients with type I diabetes mellitus. N Engl J Med 315:920–924
Dogan Y, Akarsu S, Ustundag B, Yilmaz E, Gurgoze MK (2006) Serum IL-1beta, IL-2, and Il-6 in insulin-dependent diabetic children. Mediat Inflamm 1:59206
Hussain MJ, Peakman M, Gallati H, Lo SS, Hawa M, Viberti GC et al (1996) Elevated serum levels of macrophage-derived cytokines precede and accompany the onset of IDDM. Diabetologia 39:60–69
Zier KS, Leo MM, Spielman RS, Baker L (1984) Decreased synthesis of interleukin-2 (IL-2) in insulin-dependent diabetes mellitus. Diabetes 33:552–555
Shah U, Karch L, Baker L, Zier KS (1991) Low interleukin-2 synthesis by type 1 diabetics is regulated at the pretranslational level. Clin Immunol Immunopathol 61:177–190
Lorini R, Montagna D, Lanfranchi A, Cortona L, Livieri C, Larizza D et al (1989) Alterations of in vitro interleukin 1 and 2 in diabetic children. Eur J Pediatr 148:732–734
Rapoport MJ, Mor A, Vardi P, Ramot Y, Winker R, Hindi A et al (1998) Decreased secretion of Th2 cytokines precedes up-regulated and delayed secretion of Th1 cytokines in activated peripheral blood mononuclear cells from patients with insulin-dependent diabetes mellitus. J Autoimmun 11:635–642
Berkovic MC, Jokic M, Marout J, Radosevic S, Zjacic-Rotkvic V, Kapitanovic S (2012) IL-2 -330 T/G SNP and serum values-potential new tumor markers in neuroendocrine tumors of the gastrointestinal tract and pancreas (GEP-NETs). J Mol Med (Berl) 88:423–429
Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C et al (2009) IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet 5:e1000322
Acknowledgments
We would like to thank Dr. Maria Lewandowska-Stachowiak for her excellent assistance with serum interleukin-2 measurements. We are grateful to the authorities and employees of the Regional Blood Transfusion Centre in Poznan for invaluable help with control sample collection. The study was founded by the Ministry of Science and Higher Education (Poland) under grants N402 359738 and N402 547540.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Fichna, M., Żurawek, M., Fichna, P. et al. Polymorphic variant at the IL2 region is associated with type 1 diabetes and may affect serum levels of interleukin-2. Mol Biol Rep 40, 6957–6963 (2013). https://doi.org/10.1007/s11033-013-2815-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2815-9