Abstract
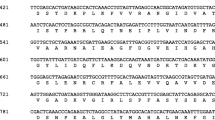
Thioredoxins (Trxs) are small ubiquitous proteins which play a regulatory role in a variety of cellular processes. In contrast to other organisms, plants have a great number of Trx types, consisting of six well-defined groups: f, m, x, and y in chloroplasts, o in mitochondria, and h mainly in cytosol. A full-length cDNA, designated VvCxxS2, encoding Trx h polypeptide was isolated and cloned from grape (Vitis vinifera L. cv. Askari) berries organ by reverse transcription polymerase chain reaction (RT-PCR). The cDNA was 381 bp nucleotides in length with a deduced amino acid of 126 residues, possessing a WCIPS active site, which belongs to the subgroup III of h-type Trxs based on phylogenetic analysis. The calculated molecular mass and the predicted isoelectric point of the deduced polypeptide are 14.25 kDa and 4.68, respectively. Nucleotide sequence analysis of genomic DNA fragment of VvCxxS2 gene revealed that this gene possesses two introns at positions identical to the previously sequenced Trx h genes. A modeling analysis indicated that VvCxxS2 shares a common structure with other Trxs, and is preferably reduced by Grx rather than NADPH-dependent thioredoxin reductase (NTR). The deduced protein sequence showed a high similarity to Trx h from other plants, in particular from castor bean (Ricinus communis), Betula pendula and sweet orange (Citrus sinensis). Semiquantitative RT-PCR experiments indicated that the transcripts of VvCxxS2 gene are present in all plant organs and different developmental stages. In addition, the higher expression of the VvCxxS2 gene was observed in berry organ as compared to the other organs.




Similar content being viewed by others
Abbreviations
- dpa:
-
Days post anthesis
- EST:
-
Expressed sequence tag
- FTR:
-
Ferredoxin-dependent thioredoxin reductase
- Grx:
-
Glutaredoxin
- GSH:
-
Glutathione
- NTR:
-
NADPH-dependent thioredoxin reductase
- ORF:
-
Open reading frame
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- Trx:
-
Thioredoxin
- UTR:
-
Untranslated region
References
Alkhalfioui F, Renard M, Frendo P, Keichinger C, Meyer Y, Gelhaye E et al (2008) A novel type of thioredoxin dedicated to symbiosis in legumes. Plant Physiol 148:424–435
Andrusier N, Nussinov R, Wolfson HJ (2007) FireDock: fast interaction refinement in molecular docking. Proteins 69:139–159
Arner ES, Holmgren A (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267:6102–6109
Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. http://nar.oxfordjournals.org/content/38/suppl_2/W529.abstract
Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18:298–305
Besse I, Buchanan BB (1997) Thioredoxin-linked plant and animal processes: the new generation. Bot Bull Acad Sin 38:1–11
Briesemeister S, Rahnenführer J, Kohlbacher O (2010) Going from where to why-interpretable prediction of protein subcellular localization. Bioinformatics 26:1232–1238
Brugidou C, Marty I, Chartier Y, Meyer Y (1992) The Nicotiana tabacum genome encodes two cytoplasmic thioredoxin genes which are differently expressed. Mol Gen Genet 238:285–293
Buchanan BB, Schürmann P, Wolosiuk RA, Jacquot JP (2002) The ferredoxin/thioredoxin system: from discovery to molecular structures and beyond. Photosynth Res 73:215–222
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc 2:953–971. doi:10.1038/nprot.2007.131
Felsenstein J (1985) Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gallie DR (1993) Post-transcriptional regulation of gene expression in plants. Ann Rev Plant Physiol Plant Mol Biol 44:77–105
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD et al (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (ed) The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Gautier MF, Lullien-Pellerin V, de Lamotte-Guery F, Guirao A, Joudrier P (1998) Characterization of wheat thioredoxin h cDNA and production of an active Triticum aestivum protein in Escherichia coli. Eur J Biochem 252:314–2324
Gelhaye E, Rouhier N, Laurent P, Sautiere PE, Martin F, Jacquot JP (2002) Isolation and characterization of an extended thioredoxin h from poplar. Physiol Plant 114:165–171
Gelhaye E, Rouhier N, Jacquot JP (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett 555:443–448
Gelhaye E, Rouhier N, Vlamis-Gardikas A, Girardet JM, Sautière PE, Sayzet M et al (2003) Identification and characterization of a third thioredoxin h in poplar. Plant Physiol Biochem 41:629–635
Gelhaye E, Rouhier N, Jacquot JP (2004) The thioredoxin h system of higher plants. Plant Physiol Biochem 42:265–271
Gelhaye E, Rouhier N, Gerard J, Jolivet Y, Gualberto J, Navrot N et al (2004) A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci USA 101:14545–14550
Gelhaye E, Rouhier N, Navrot N, Jacquot JP (2005) The plant thioredoxin system. Cell Mol Life Sci 62:24–35
Geourjon C, Deleage G (1995) SOPMA: significant improvement in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11:681–684
Heidari Japelaghi R, Haddad R, Garoosi GA (2011) Rapid and efficient isolation of high quality nucleic acids from plant tissues rich in polyphenols and polysaccharides. Mol Biotechnol. doi:10.1007/s12033-011-9384-8
Ishiwatari Y, Honda C, Kawashima I, Nakamura S, Hirano H, Mori S et al (1995) Thioredoxin h is one of the major proteins in rice phloem sap. Planta 195:456–463
Jacquot JP, Lancelin JM, Meyer Y (1997) Thioredoxin: structure and function in plant cells. New Phytol 136:543–570
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jiang H, Song W, Li A, Yang X, Sun D (2011) Identification of genes differentially expressed in cauliflower associated with resistance to Xanthomonas campestris pv. Campestris. Mol Biol Rep 38:621–629
Juttner J, Olde D, Langridge P, Baumann U (2000) Cloning and expression of a distinct cluster of plant thioredoxins. Eur J Biochem 267:7109–7117
Kim YJ, Shim JS, Krishna PR, Kim SY, In JG, Kim MK et al (2008) Isolation and characterization of a glutaredoxin gene from Panax ginseng C. A. Meyer. Plant Mol Biol Rep 26:335–349
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T et al (2005) ConSurf: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acid Res. http://www.ncbi.nlm.nih.gov/pubmed/15980475
Lennon BW, Williams CH, Ludwig ML (2000) Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science 289:1190–1194
Li HL, Lu HZ, Guo D, Tian WM, Peng SQ (2010) Molecular characterization of a thioredoxin h gene (HbTRX1) from Hevea brasiliensis showing differential expression in latex between self-rooting juvenile clones and donor clones. Mol Biol Rep 38:1989–1994
Maeda K, Hagglund P, Finnie C, Svensson B, Henriksen A (2008) Crystal structures of barley thioredoxin h isoforms HvTrxh1 and HvTrxh2 reveal features involved in protein recognition and possibly in discriminating the isoform specificity. Protein Sci 17:1015–1024
Meng L, Wong JH, Feldman LJ, Lemaux PG, Buchanan BB (2010) A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc Natl Acad Sci USA 107:3900–3905
Messing J, Geraghty D, Heidecker G, Hu NT, Kridl J, Rubenstein I (1983) Plant gene structure. In: Kosuge T, Meredith CP, Hollaender A (eds) Genetic engineering of plants. Plenum Press, New York, pp 211–227
Meyer Y, Vignols F, Reichheld JP (2002) Classification of plant thioredoxins by sequence similarity and intron position. Method Enzymol 347:394–402
Meyer Y, Siala W, Bashandy T, Riondet C, Vignols F, Reichheld JP (2008) Glutaredoxins and thioredoxins in plants. Biochim Biophys Acta 1783:589–600
Montrichard F, Renard M, Alkhalfioui F, Duval FD, Macherel D (2003) Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiol 132:1707–1715
Nimrod G, Schushan M, Steinberg DM, Ben-Tal N (2008) Detection of functionally important regions in hypothetical proteins of known structure. Structure 16:1755–1763
Qiu L, Ma Z, Jiang S, Wang W, Zhou F, Huang J et al (2009) Molecular cloning and mRNA expression of peroxiredoxin gene in black tiger shrimp (Penaeus monodon). Mol Biol Rep 37:2821–2827
Rouhier N, Gelhaye E, Jacquot JP (2002) Redox control by dithiol-disulfide exchange in plants. Part II. Cytosolic and mitochondrial systems. Ann NY Acad Sci 973:520–528
Rouhier N, Gelhaye E, Jacquot JP (2004) Plant glutaredoxins: still mysterious reducing systems. Cell Mol Life Sci 61:1266–1277
Russell RB, Breed J, Barton GJ (1992) Conservation analysis and secondary structure prediction of the SH2 family of phosphotyrosine binding domains. FEBS Lett 304:15–20
Sahrawy M, Hecht V, Lopez-Jaramillo J, Chueca A, Chartier Y, Meyer Y (1996) Intron position as an evolutionary marker of thioredoxins and thioredoxin domains. J Mol Evol 42:422–431
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acid Res. http://www.ncbi.nlm.nih.gov/pubmed/15980490
Schobert C, Baker L, Szederkenyi J, Grossmann P, Komor E, Hayashi H (1998) Identification of immunologically related proteins in sieve-tube exudate collected from monocotyledonous and dicotyledonous plants. Planta 206:245–252
Schröter Y, Steiner S, Matthäi K, Pfannschmidt T (2010) Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 10:2191–2204
Serrato AJ, Cejudo FJ (2003) Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta 217:392–399
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40
Acknowledgments
We wish to thank Dr. Ramin Hosseini and Dr. A. H. Beiki (Imam Khomeini International University, Qazvin, IR of Iran) for helpful and technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Japelaghi, R.H., Haddad, R. & Garoosi, GA. Isolation, identification and sequence analysis of a thioredoxin h gene, a member of subgroup III of h-type Trxs from grape (Vitis vinifera L. cv. Askari). Mol Biol Rep 39, 3683–3693 (2012). https://doi.org/10.1007/s11033-011-1143-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1143-1