Abstract
The cDNA code of thioredoxin h, designated as HbTRX1, was isolated from Hevea brasiliensis by rapid amplification of cDNA ends. HbTRX1 contained a 542-bp open reading frame encoding 123 amino acids. The deduced HbTRX1 protein showing high identity to thioredoxin h of other plant species was predicted to possess the conserved catalytic site WCXPC. Semiquantitative reverse transcription-polymerase chain reaction analysis revealed that HbTRX1 was constitutively expressed in all tested tissues. HbTRX1 transcripts accumulated at relatively low levels in the flower, somatic embryo, and leaves, while HbTRX1 transcripts accumulated at relatively high levels in the callus and latex. The HbTRX1 transcript was expressed at different levels, with higher levels in self-rooting juvenile clones than in their donor clones. HbTRX1 was expressed in Escherichia coli, and its activity was demonstrated using the dithiothreitol-dependent insulin assay. This work provides a basis for studying the biological function of thioredoxin h in rubber tree.



Similar content being viewed by others
Abbreviations
- ORF:
-
Open reading frame
- RACE:
-
Rapid amplification of cDNA ends
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- UTR:
-
Untranslated region
References
Clément-Demange A, Priyadarshan PM, Hoa TTT, Venkatachalam P (2007) Hevea rubber breeding and genetics. In: Janick J (ed) Plant breeding reviews, vol 29. Wiley, New York, pp 177–281
Chandrashekar TR, Mydin KK, Alice J, Varghese YA, Saraswathyamma CK (1997) Intraclonal variability for yield in rubber (Hevea brasiliensis). Ind J Nat Rubber Res 10:43–47
Carron MP, Enjalric F, Lardet L, Deschamps A (1989) Rubber (Hevea brasiliensis Muell. Arg.). In: Bajaj (ed) Biotechnology in agriculture and forestry, vol 5. Springer-Verlag, Germany, pp 222–245
Chen XT, Wang ZY, Wu HD, Zhang XJ (2001) Selection of optimum planting material of Hevea brasiliensis: Self-rooting juvenile clone. In: Sainte-Beuve J (ed) Annual IRRDB Meeting CIRAD. Montpellier, France
Wang Z, Zeng X, Chen C, Wu H, Li Q, Fan G, Lu W (1980) Induction of rubber plantlets from anther of Hevea brasiliensis in vitro. Chin J Trop Crops 1:25–26
Yang SQ, Mo YY (1994) Some physiological properties of latex from somatic plants of Hevea brasiliensis. Chin J Trop Crops 15:13–20
Hao BZ, Wu JL (1996) The high-yield characteristics of rubber tree juvenile clone. Trop Agric Sci 2:1–8
Liang XL, Li HL, Peng SQ (2009) Cloning and expression of HbTCTP from Hevea brasiliensis. Mol Plant Breed 7:194–198
Chow KS, Wan KL, Mat IMN, Bahari A, Tan SH, Harikrishna K, Yeang HY (2007) Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J Exp Bot 58:2429–2440
Venkatachalam P, Thulaseedharan A, Raghothama K (2007) Identification of expression profiles of tapping panel dryness (TPD) associated genes from the latex of rubber tree (Hevea brasiliensis Muell. Arg). Planta 226:499–515
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation version II. Plant Mol Biol Rep 1:19–21
Tang C, Qi J, Li H, Zhang C, Wang Y (2007) A convenient and efficient protocol for isolating high-quality RNA from latex of Hevea brasiliensis (para rubber tree). J Biochem Biophys Methods 70:749–754
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 24:4876–4882
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Holmgren A (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254:9627–9632
Schürmann P, Jacquot JP (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51:371–400
Gelhaye E, Rouhier N, Jacquot JP (2004) The thioredoxin h system of higher plants. Plant Physiol Biochem 42:265–271
Reicheld JP, Mestres-Ortega D, Laloi C, Meyer Y (2002) The multigenic family of thioredoxin h in Arabidopsis thaliana: specific expression and stress response. Plant Physiol Biochem 40:685–690
Cazalis R, Pulido P, Aussenac T, Pérez-Ruiz JM, Cejudo FJ (2006) Cloning and characterization of three thioredoxin h isoforms from wheat showing differential expression in seeds. J Exp Bot 57:2165–2172
Wong JH, Kim YB, Ren PH, Cai N, Cho MJ, Hedden P, Lemaux PG, Buchanan BB (2002) Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA 99:16325–16330
Shahpiri A, Svensson B, Finnie C (2008) The NADPH-dependent thioredoxin reductase/thioredoxin system in germinating barley seeds: gene expression, protein profiles, and interactions between isoforms of thioredoxin h and thioredoxin reductase. Plant Physiol 146:789–799
Gelhaye E, Rouhier N, Jacquot JP (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett 555:443–448
Jung BG, Lee KO, Lee SS, Chi YH, Jang HH, Kang SS, Lee K, Lim D, Yoon SC, Yun DJ, Inoue Y, Cho MJ, Lee SY (2002) A Chinese cabbage cDNA with high sequence identity to phospholipids hydroperoxide glutathione peroxidases encodes a novel isoform of thioredoxin-dependent peroxidase. J Biol Chem 277:12572–12578
Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fay E, Meyer Y, Jacquot JP (2001) Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol 127:1299–1309
Park SK, Jung YJ, Lee JR, Lee YM, Jang HH, Lee SS, Park JH, Kim SY, Moon JC, Lee SY, Chae HB, Shin MR, Jung JH, Kim MG, Kim WY, Yun DJ, Lee KO, Lee SY (2009) Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol 150:552–561
Pulido P, Cazalis R, Cejudo FJ (2009) An antioxidant redox system in the nucleus of wheat seed cells suffering oxidative stress. Plant J 57:132–145
Baumann U, Juttner J (2002) Plant thioredoxins: the multiplicity conundrum. Cell Mol Life Sci 59:1042–1057
Maeda K, Finnie C, Svensson B (2003) Cy5 maleimide-labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. Biochem J 24:497–507
Wong JH, Balmer Y, Cai N, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2003) Unraveling thioredoxin-linked metabolic processes of cereal starchy endosperm using proteomics. FEBS Lett 547:151–156
Yano H, Wong JH, Lee YM, Cho MJ, Buchanan BB (2001) A strategy for the identification of proteins targeted by thioredoxin. Proc Natl Acad Sci USA 98:4794–4799
Kush A (1994) Isoprenoid biosynthesis: the Hevea factory. Plant Physiol Biochem 32:761–767
Auzac J (1989) Factors involved in the stopping of latex flow after tapping. In: d’Auzac J, Chrestin (eds) Physiology of the rubber tree latex. CRC Press, Inc., Boca Raton, pp 257–280
Chrestin H, Gidrol X, Kush A (1997) Towards a latex molecular diagnostic of yield potential and the genetic engineering of the rubber tree. Euphytica 96:77–82
Acknowledgments
This work was supported by the National Nonprofit Institute Research Grant of CATAS-ITBB (ITBBZD0712) and the National Natural Science Foundation of China (30960307). We deeply thank Professor Xiong-Ting Chen (Institute of Tropical Biosciences and Biotechnology, Chinese Academy of Tropical Agricultural Sciences) for his collaboration in the treatment of plant materials.
Author information
Authors and Affiliations
Corresponding author
Additional information
Hui-Liang Li and Hui-Zhong Lu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2010_321_MOESM1_ESM.tif
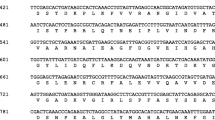
Supplementary 1 Sequence alignment of the deduced HbTRX1 and other plant thioredoxins h. VvTRX (Vitis vinifera, ABX79345), ClTRX (Codonopsis lanceolata, BAE16559), GmTRX (Glycine max, ABV71991), HbTRX (H. brasiliensis, AF133127.1), PtTRX (Populus tremula × Populus, AF483266), ATTRX1 (A. thaliana, P29448), ATTRX2 (A. thaliana, NM_123358), ATTRX3 (A. thaliana, NM_123664), ATTRX4 (A. thaliana, NM_101829), ATTRX5 (A. thaliana, NM_103588), ATTRX6 (A. thaliana, AAD39316), ATTRX7 (A. thaliana, AAG52561), and ATTRX8 (A. thaliana, AAG51342). Amino acid residues that are identical in all six sequences are shaded in black, and well-conserved residues are shaded in gray. (TIFF 1143 kb)
11033_2010_321_MOESM2_ESM.tif
Supplementary 2 Phylogenetic analysis of HbTRX1 and other species by MEGA version 3.1 from CLUSTAL W alignments. The thioredoxins h used in the evolutionary analysis are retrieved from the GenBank, including VvTRX (V. vinifera, ABX79345), ClTRX (C. lanceolata, BAE16559), GmTRX (G. max, ABV71991), HbTRX (H. brasiliensis, AF133127.1), PtTRX (P. tremula × Populus, AF483266), ATTRX1 (A. thaliana, P29448), ATTRX2 (A. thaliana, NM_123358), ATTRX3 (A. thaliana, NM_123664), ATTRX4 (A. thaliana, NM_101829), ATTRX5 (A. thaliana, NM_103588), ATTRX6 (A. thaliana, AAD39316), ATTRX7 (A. thaliana, AAG52561), and ATTRX8 (A. thaliana, AAG51342). (TIFF 372 kb)
11033_2010_321_MOESM3_ESM.tif
Supplementary 3 Nucleotide sequence of the promoter region of HbTRX1. The start code is boxed. The cis-acting regulatory elements are underlined. (TIFF 406 kb)
Rights and permissions
About this article
Cite this article
Li, HL., Lu, HZ., Guo, D. et al. Molecular characterization of a thioredoxin h gene (HbTRX1) from Hevea brasiliensis showing differential expression in latex between self-rooting juvenile clones and donor clones. Mol Biol Rep 38, 1989–1994 (2011). https://doi.org/10.1007/s11033-010-0321-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0321-x