Abstract
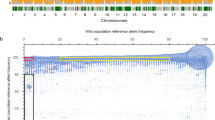
To discover new SNPs and develop an easy assay method in soybean, we compared the high-throughput pyrosequencing ESTs with whole genome sequences in different soybean varieties and identified 3899 SNPs. Transitions were found to be much more frequent than transversions in these SNPs. We found that SNPs were widely distributed in the soybean genome, targeting numerous genes involved in various physiological and biochemical processes influencing important agronomic traits. A set of 16 SNPs were validated in nine soybean varieties, and seven SNPs were converted into CAPS. From functional gene association analysis, the marker CAPS282 on the 3′-UTR of gene Glyma07g03490 was identified as associated with 100-seed weight in soybean. The SNP discovery and CAPS markers conversion system developed in this study is fast and cost effective, and holds great promise for molecular-assisted breeding of soybean.


Similar content being viewed by others
References
Agarwal M, Shrivastava N, Padh H (2008) Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep 27(4):617–631
Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok P-Y, Gish WR (1999) A general approach to single-nucleotide polymorphism discovery. Nat Genet 23(4):452–456
Batley J, Barker G, O’Sullivan H, Edwards KJ, Edwards D (2003) Mining for single nucleotide polymorphisms and insertions/deletions in maize expressed sequence tag data. Plant Physiol 132(1):84–91
Kota R, Varshney R, Prasad M, Zhang H, Stein N, Graner A (2008) EST-derived single nucleotide polymorphism markers for assembling genetic and physical maps of the barley genome. Functional & Integrative Genomics 8(3):223–233
Ganal MW, Altmann T, Roder MS (2009) SNP identification in crop plants. Curr Opin Plant Biol 12(2):211–217
Shendure J, Ji H (2008) Next-generation DNA sequencing. Nat Biotech 26(10):1135–1145
Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS (2007) SNP discovery via 454 transcriptome sequencing. Plant J 51(5):910–918
Ardashir KM, Daniel LEW, Russel FR, Robert JH (2009) A high-throughput assay for rapid and simultaneous analysis of perfect markers for important quality and agronomic traits in rice using multiplexed MALDI-TOF mass spectrometry. Plant Biotechnol J 7(4):355–363
Aurélie B, Marie Christine Le P, Mireille D, Florence E-V, Isabelle B, Alberto C, Annabelle H, Dominique B, Catherine R (2009) High-throughput single nucleotide polymorphism genotyping in wheat (Triticum spp.). Plant Biotechnol J 7(4):364–374
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung J, Specht JE (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39(5):1464–1490
Song Q, Marek L, Shoemaker R, Lark K, Concibido V, Delannay X, Specht J, Cregan P (2004) A new integrated genetic linkage map of the soybean. TAG Theor Appl Genet 109(1):122–128
Ning Z, Caccamo M, and Mullikin JC (2005) ssahaSNP-a polymorphism detection tool on a whole genome scale. In: Proceedings of the 2005 IEEE computational systems bioinformatics conference—workshops, IEEE Computer Society, Washington, DC, USA, pp. 251–254
Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, Radenbaugh A, Singh S, Swing V, Tissier C, Zhang P, and Huala E (2007) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucl Acids Res. gkm965
Mewes HW, Frishman D, Mayer KFX, Munsterkotter M, Noubibou O, Pagel P, Rattei T, Oesterheld M, Ruepp A, Stumpflen V (2006) MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucl Acids Res 34(suppl_1):D169–D172
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25(17):3389–3402
Ren G, Chen H, Zhang LZ, Lan XY, Wei TB, Li MJ, Jing YJ, Lei CZ, Wang JQ (2010) A coding SNP of LHX4 gene is associated with body weight and body length in bovine. Mol Biol Rep 37(1):417–422
Choi I-Y, Hyten DL, Matukumalli LK, Song Q, Chaky JM, Quigley CV, Chase K, Lark KG, Reiter RS, Yoon M-S, Hwang E-Y, Yi S-I, Young ND, Shoemaker RC, van Tassell CP, Specht JE, Cregan PB (2007) A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics 176(1):685–696
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single-nucleotide polymorphisms in soybean. Genetics 163(3):1123–1134
Michael I, Chris D, Jacqueline B, David E (2009) Discovering genetic polymorphisms in next-generation sequencing data. Plant Biotechnol J 7(4):312–317
Lai X, Lan X, Chen H, Wang X, Wang K, Wang M, Yu H, Zhao M (2009) A novel SNP of the Hesx1 gene in bovine and its associations with average daily gain. Mol Biol Rep 36(7):1677–1681
He X, Xu X, Liu B (2009) Molecular characterization, chromosomal localization and association analysis with back-fat thickness of porcine LPIN2 and LPIN3. Mol Biol Rep 36(7):1819–1824
Niu PX, Huang Z, Li CC, Fan B, Li K, Liu B, Yu M, Zhao SH (2009) Cloning, chromosomal localization, SNP detection and association analysis of the porcine IRS-1 gene. Mol Biol Rep 36(8):2087–2092
He X, Gao H, Liu C, Fan B, and Liu B (2010) Cloning, chromosomal localization, expression profile and association analysis of the porcine WNT10B gene with backfat thickness. Mol Biol Rep. doi:10.1007/s11033-010-9978-4
Li Y, Yang SL, Tang ZL, Cui WT, Mu YL, Chu MX, Zhao SH, Wu ZF, Li K, Peng KM (2010) Expression and SNP association analysis of porcine FBXL4 gene. Mol Biol Rep 37(1):579–585
Xu X, Qiu H, Du ZQ, Fan B, Rothschild MF, Yuan F, Liu B (2010) Porcine CSRP3: polymorphism and association analyses with meat quality traits and comparative analyses with CSRP1 and CSRP2. Mol Biol Rep 37(1):451–459
Cao GL, Chu MX, Fang L, Di R, Feng T, and Li N (2010) Analysis on DNA sequence of KiSS-1 gene and its association with litter size in goats. Mol Biol Rep. doi:10.1007/s11033-010-0049-7
Acknowledgments
Soybean genome sequence data were produced by the U.S. Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the user community, and we thank them. The project was supported by the National High-Technology Program (2006AA100104, 200008AA10Z153, 2009ZX08009-032B), the Key Research Plan of Heilongjiang Province (GA06B103), and the Innovation Research Group of NEAU (CXT004).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yongjun Shu and Yong Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shu, Y., Li, Y., Zhu, Z. et al. SNPs discovery and CAPS marker conversion in soybean. Mol Biol Rep 38, 1841–1846 (2011). https://doi.org/10.1007/s11033-010-0300-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0300-2