Abstract
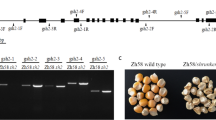
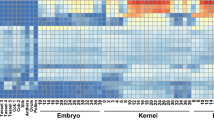
The discovery and characterization of the opaque endosperm gene provide ideas and resources for the production and application of maize. We found an o213 mutant whose phenotype was opaque and shrunken endosperm with semi-dwarf plant height. The protein, lipid, and starch contents in the o213 endosperm were significantly decreased, while the free amino acid content in the o213 endosperm significantly increased. The aspartic acid, asparagine, and lysine contents were raised in the o213 endosperm by 6.5-, 8.5-, and 1.7-fold, respectively. Genetic analysis showed that this o213 mutant is a recessive single-gene mutation. The position mapping indicated that o213 is located in a 468-kb region that contains 11 protein-encoding genes on the long arm of chromosome 5. The coding sequence analysis of candidate genes between the WT and o213 showed that ZmYSL2 had only a single-base substitution (A–G) in the fifth exon, which caused methionine substitution to valine. Sequence analysis and the allelic test showed that o213 is a new mutant allele of ZmYSL2. The qRT-PCR results indicated that o213 is highly expressed in the stalks and anthers. Subcellular localization studies showed that o213 is a membrane transporter. In the variation analysis of o213, the amplification of 65 inbred lines in GWAS showed that this 3-bp deletion of the first exon of o213 was found only in temperate inbred lines, implying that the gene was artificially affected in the selection process. Our results suggest that o213 is an important endosperm development gene and may serve as a genetic resource.






Similar content being viewed by others
References
Albenne C, Canut H, Boudart G, Zhang Y, San Clemente H, Pont-Lezica R, Jamet E (2009) Plant cell wall proteomics: mass spectrometry data, a trove for research on protein structure/function relationships. Mol Plant 2:977–989. https://doi.org/10.1093/mp/ssp059
Bhatnagar S, Betrán F, Rooney L (2004) Combining abilities of quality protein maize inbreds. Crop Science - CROP SCI 44(6):1997–2005. https://doi.org/10.2135/cropsci2004.1997
Bogs J, Bourbouloux A, Cagnac O, Wachter A, Rausch T, Delrot S (2003) Functional characterization and expression analysis of a glutathione transporter, BjGT1, from Brassica juncea: evidence for regulation by heavy metal exposure. Plant, Cell Environ 26:1703–1711. https://doi.org/10.1046/j.1365-3040.2003.01088.x
Boi M, Porceddu M, Cappai G, Giudici G, Bacchetta G (2020) Effects of zinc and lead on seed germination of Helichrysum microphyllum subsp. tyrrhenicum, a metal-tolerant plant. Int J Environ Sci Technol 17:1917–1928. https://doi.org/10.1007/s13762-019-02589-9
Chen F, Zhu SW, Xiang Y, Jiang Y, Cheng BJ (2010) Molecular marker-assisted selection of the ae alleles in maize. Genetics and Molecular Research : GMR 9:1074–1084. https://doi.org/10.4238/vol9-2gmr799
Curie C, Panaviene Z, Loulergue C, Dellaporta S, Briat J-F, Walker E (2001) Maize Yellow Stripe 1 (YS 1) encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346–349. https://doi.org/10.1038/35053080
Dangol S, Singh R, Chen Y, Jwa NS (2017) Visualization of multicolored in vivo organelle markers for co-localization studies in Oryza sativa. Mol. Cells 40(11):828–836. https://doi.org/10.14348/molcells.2017.0045
Delrot S, Atanassova R, Maurousset L (2000) Regulation of sugar, amino acid and peptide plant membrane transporters. Biochem Biophys Acta 1465:281–306. https://doi.org/10.1016/S0005-2736(00)00145-0
Deng Q, Shi-Quan W, Al-Ping Z, Hong-Yu Z, Ping L (2007) Breeding rice restorer lines with high resistance to bacterial blight by using molecular marker-assisted selection. Int Rice Res Notes 32
Ester R, Ang J (2021) Embryo and endosperm function and failure in Solanum species and hybrids.
Falchuk KH, Hardy C, Ulpino L, Vallee BL (1978) RNA metabolism, manganese, and RNA polymerases of zinc-sufficient and zinc-deficient Euglena gracilis. Proc Natl Acad Sci 75(9):4175. https://doi.org/10.1073/pnas.75.9.4175
Feng F, Qi W, Lv Y, Yan S, Xu L, Yang W, Yuan Y, Chen Y, Zhao H, Song R (2018) OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell 30(2):375–396. https://doi.org/10.1105/tpc.17.00616
Gupta H, Pk A, Mahajan V, Gs B, A K, P V, A S, Saha S, Babu R, Mc P, Vp M, (2009) Quality protein maize for nutritional security: rapid development of short duration hybrids through molecular marker assisted breeding. Curr Sci 96:230–237
He Y, Yang Q, Yang J, Wang Y-F, Sun X, Wang S, Qi W, Ma Z, Song R (2021) shrunken4 is a mutant allele of ZmYSL2 that affects aleurone development and starch synthesis in maize. Genetics. https://doi.org/10.1093/genetics/iyab070
Janick J (2021) New crops and the search for new food resources.
Jin X, Fu Z, Ding D, Li W, Liu Z, Tang J (2013) Proteomic identification of genes associated with maize grain-filling rate. PLoS ONE 8:e59353. https://doi.org/10.1371/journal.pone.0059353
Kiesselbach TA (1980) The structure and reproduction of corn. Research Bulletin 161:1–96
Letchworth M, Lambert R (1998) Pollen parent effects on oil, protein, and starch concentration in maize kernels. Crop Science - CROP SCI 38(2):363–367. https://doi.org/10.2135/cropsci1998.0011183X003800020015x
Lubkowitz M (2006) The OPT family functions in long-distance peptide and metal transport in plants. Genet Eng 27:35–55. https://doi.org/10.1007/0-387-25856-6_3
Lubkowitz M, Hauser L, Breslav MF, Naider F, Becker J (1997) An oligo transport gene from Candida albicans. Microbiology (reading, England) 143(Pt 2):387–396. https://doi.org/10.1099/00221287-143-2-387
Massey L (2003) Dietary animal and plant protein and human bone health: a whole foods approach. J Nutr 133:862S-865S. https://doi.org/10.1093/jn/133.3.862S
Mertz E, Bates LSET, Nelson O (1964) Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science (new York, NY) 145:279–280. https://doi.org/10.1126/science.145.3629.279
Myers AM, James MG, Lin Q, Yi G, Stinard PS, Hennen-Bierwagen TA, Becraft PW (2011) Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23(6):2331–2347. https://doi.org/10.1105/tpc.111.087205
Olsen O-A (2020) The modular control of cereal endosperm development. Trends Plant Sci 25(3):279–290. https://doi.org/10.1016/j.tplants.2019.12.003
Ozturk L, Yazici A, Yucel C, Alkantorun A, Çekiç C, Bağcı S, Ozkan H, Braun HJ, Sayers Z, Cakmak I (2006) Concentration and localization of zinc during seed development and germination in wheat. Physiol Plant 128:144–152. https://doi.org/10.1111/j.1399-3054.2006.00737.x
Sabelli PA, Larkins BA (2009) The development of endosperm in grasses. Plant Physiol 149(1):14–26. https://doi.org/10.1104/pp.108.129437
Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, von Wiren N (2004) ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J Biol Chem 279(10):9091–9096. https://doi.org/10.1074/jbc.M311799200
Schmidt R, Burr F, Burr B (1987) Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science (new York, NY) 238:960–963. https://doi.org/10.1126/science.2823388
Segal G, Song R, Messing J (2003) A new opaque variant of maize by a single dominant RNAi-inducing transgene. Genetics 165:387–397. https://doi.org/10.1093/genetics/165.1.387
Sene M, Thevenot C, Prioul JL (1997) Simultaneous spectrophotometric determination of amylose and amylopectin in starch from maize kernel by multi-wavelength analysis. J Cereal Sci 26(2):211–221. https://doi.org/10.1006/jcrs.1997.0124
Snehi Dwivedi R, Takkar PN (1974) Ribonuclease activity as an index of hidden hunger of zinc in crops. Plant Soil 40(1):173–181. https://doi.org/10.1007/BF00011420
Stacey MG, Koh S, Becker J, Stacey G (2002) AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell 14(11):2799–2811. https://doi.org/10.1105/tpc.005629
Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G (2006) Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta 223(2):291–305. https://doi.org/10.1007/s00425-005-0087-x
Tunes L, Muniz M, Torales-Salinas J, Suarez-Castellanos C, Lemes E (2015) Response of wheat (Triticum aestivum L.) seedlings to seed zinc application. Agrociencia 49:623–636
Ullah A, Farooq M, Nadeem F, Rehman A, Nawaz A, Naveed M, Wakeel A, Hussain M (2020) Zinc seed treatments improve productivity, quality and grain biofortification of desi and kabuli chickpea (Cicer arietinum). Crop Pasture Sci 71:725–738. https://doi.org/10.1071/CP19266
Vasal SK (2000) High quality protein corn. In. pp 85–129
von Wirén N, Klair S, Bansal S, Briat JF, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both Fe<sup>III</sup> and Fe<sup>II</sup> Implications for metal transport in plants. Plant Physiology 119(3):1107. https://doi.org/10.1104/pp.119.3.1107
von Wiren N, Marschner H, Romheld V (1996) Roots of iron-efficient maize also absorb phytosiderophore-chelated zinc. Plant Physiol 111(4):1119. https://doi.org/10.1104/pp.111.4.1119
von Wiren N, Mori S, Marschner H, Romheld V (1994) Iron inefficiency in maize mutant ys1 (Zea mays L. cv Yellow-Stripe) is caused by a defect in uptake of iron phytosiderophores. Plant Physiology 106 (1):71. https://doi.org/10.1104/pp.106.1.71
Wang C, Kovacs M (2002) Swelling index of glutenin test. II. Application in prediction of dough properties and end-use quality 1. Cereal Chemistry - CEREAL CHEM 79:190–196. https://doi.org/10.1094/CCHEM.2002.79.2.190
Wang G, Sun X, Wang G, Wang F, Gao Q, Sun X, Tang Y, Chang C, Lai J, Zhu L, Xu Z, Song R (2011) Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 189(4):1281–1295. https://doi.org/10.1534/genetics.111.133967
Wang G, Wang F, Wang G, Wang F, Zhang X, Zhong M, Zhang J, Lin D, Tang Y, Xu Z, Song R (2012) Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 24(8):3447–3462. https://doi.org/10.1105/tpc.112.101360
Williams L, Miller A (2001) Transporters responsible for the uptake and partitioning of nitrogenous solutes. Annu Rev Plant Physiol Plant Mol Biol 52:659–688. https://doi.org/10.1146/annurev.arplant.52.1.659
Wu Y, Messing J (2014) Proteome balancing of the maize seed for higher nutritional value. Front Plant Sci 5:240. https://doi.org/10.3389/fpls.2014.00240
Yao D, Qi W, Li X, Yang Q, Yan S, Ling H, Wang G, Wang G, Song R (2016) Maize opaque10 encodes a cereal-specific protein that is essential for the proper distribution of zeins in endosperm protein bodies. PLOS Genet 12(8):e1006270. https://doi.org/10.1371/journal.pgen.1006270
Yen M-R, Tseng YH, Saier MH (2001) Maize Yellow Stripe 1, an iron-phytosiderophore uptake transporter, is a member of the oligopeptide transporter (OPT) family. Microbiology (reading, England) 147:2881–2883. https://doi.org/10.1099/00221287-147-11-2881
Zang J, Huo Y, Liu J, Zhang H, Liu J, Chen H (2020) Maize YSL2 is required for iron distribution and development in kernels. J Exp Bot 71(19):5896–5910. https://doi.org/10.1093/jxb/eraa332
Zhang MY, Bourbouloux A, Cagnac O, Srikanth CV, Rentsch D, Bachhawat AK, Delrot S (2004) A novel family of transporters mediating the transport of glutathione derivatives in plants. Plant Physiol 134(1):482–491. https://doi.org/10.1104/pp.103.030940
Zhang S, Ni D, Yi C, Li L, Wang X, Wang Z, Yang J (2005) Reducing amylose content of indica rice variety 057 by molecular marker-assisted selection. Zhongguo Shuidao Kexue 19(5):467–470
Zhang X, Zhang H, Li L, Lan H, Ren Z, Liu D, Wu L, Liu H, Jaqueth J, Li B, Pan G, Gao S (2016) Characterizing the population structure and genetic diversity of maize breeding germplasm in Southwest China using genome-wide SNP markers. BMC Genomics 17(1):697. https://doi.org/10.1186/s12864-016-3041-3
Funding
This research was funded and supported by China Agriculture Research System of MOF and MARA, and Sichuan Science and Technology Support Project (2021YFYZ0020, 2021YFYZ0027, 2021YFFZ0017).
Author information
Authors and Affiliations
Contributions
Y.W. designed the research and performed the experiment, analyzed data, and wrote the manuscript. H.H., H.Z., H.Z., Z.Z., and J.G. investigate the agronomic traits. D.L. and L.W. provide reagents and consumables. S.G. and D.G. contributed to plant materials management. X.Z. and B.L. revised the manuscript. S.G. contributed to critically reading this manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

11032_2022_1278_MOESM1_ESM.jpg
Figure S1. Comparison of the difference of metal ion concentration between WT and o213 kernels a Copper concentration of WT and o213 mutant in embryo, endosperm and seed of 18DAP and 27DAP. b Manganese concentration of WT and o213 mutant in embryo, endosperm and seed of 18DAP and 27DAP. c Zinc concentration of WT and o213 mutant in embryo, endosperm and seed of 18DAP and 27DAP. Student's t test, (*) P<0.05, (***) P<0.001, and n, not significant. (JPG 227 KB)




11032_2022_1278_MOESM5_ESM.jpg
Figure S5. Analysis of the transmembrane domain of the four genotypes, ZmYSL2 was used as a control. The red box is the deleted amino acid (JPG 862 KB)

11032_2022_1278_MOESM7_ESM.pdf
Table S1 Information on specific primers for gene mapping, cloning, qRT–PCR, subcellular localization and evolution analysis (PDF 110 KB)
11032_2022_1278_MOESM9_ESM.pdf
Table S3 Related agronomic traits and quality content data of the four genotypes, t-test among the traits of the four genotypes (PDF 23 KB)
Rights and permissions
About this article
Cite this article
Wang, Y., Zhang, X., Luo, B. et al. Identification of a new mutant allele of ZmYSL2 that regulates kernel development and nutritional quality in maize. Mol Breeding 42, 7 (2022). https://doi.org/10.1007/s11032-022-01278-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-022-01278-9