Abstract
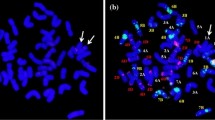
PI 554419, formerly designated as Ae. uniaristata, showed significant difference with other Ae. uniaristata and Ae. comosa accessions in morphological traits at the seedling stage and its leaf color, length, and width behaved as an intermediate type. In this study, we reclassified PI 554419 as Ae. comosa subsp. comosa by comparing the fluorescence in situ hybridization (FISH) signals and the patterns of PCR-based landmark unique gene (PLUG) markers and conserved orthologous set (COS) markers of PI 554419 with other Ae. uniaristata and Ae. comosa accessions as well as the taxonomic character of spike morphology. A disomic 1M/1D substitution line NB 4-8-5-9 derived from PI 554419 was identified from a distant hybridization of Ae. comosa with common wheat (STM 10/CSph1b//CM 39///13 P2-6) by the molecular cytological method. Furthermore, the agronomic and seed morphological traits, as well as the flour processing quality properties of NB 4-8-5-9, were compared with those of its three common wheat parents in two different locations during the 2017–2018 growing seasons. The agronomical traits of NB 4-8-5-9 were similar to or even better than its parents. The seed size-related traits of NB 4-8-5-9 were better than those of all three parents, and the 1000-grain weight and grain width were close to those of Chuanmai 39 (CM 39) and 13 P2-6 and larger than those of CSph1b. The processing quality properties of NB 4-8-5-9 were more similar to those of 13 P2-6 and CSph1b but less similar to those of CM 39. The 1M/1D substitution line NB 4-8-5-9 could further be used for developing translocation lines with 1M segment.




Similar content being viewed by others
Data availability
The seeds in the current study are available from the corresponding author upon request.
References
Abdolmalaki Z, Mirzaghaderi G, Mason AS, Badaeva ED (2019) Molecular cytogenetic analysis reveals evolutionary relationships between polyploid Aegilops species. Plant Syst Evol 305:459–475
Badaeva ED, Dedkova OS, Zoshchuk SA, Amosova AV, Reader SM, Bernard M, Zelenin AV (2011) Comparative analysis of the N-genome in diploid and polyploidy Aegilops species. Chromosom Res 19:541–548
Badaeva ED, Dedkova OS, Pukhalskiy VA, Zelenin AV (2012) Cytogenetic comparison of N-genome Aegilops L. Species. Russ J Genet 48:522–531
Cuadrado A, Cardoso M, Jouve N (2008) Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome 51:809–815
Cui F, Zhao CH, Ding AM, Li J, Wang L, Li XF, Bao YG, Li JM, Wang HG (2014) Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet 127:659–675
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fu SL, Chen L, Wang YY, Li M, Yang ZJ, Qiu L, Yan BJ, Ren ZL, Tang ZX (2015) Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep 5:10552
Garg M, Tsujimoto H, Gupta RK, Kumar A, Kaur N, Kumar R, Chunduri V, Sharma NK, Chawla M, Sharma S, Mundey JK (2016) Chromosome specific substitution lines of Aegilops geniculata alter parameters of bread making quality of wheat. PLoS One 11:e0162350
Georgiou A, Karataglis S, Roupakias D (1992) Inter- and intra plant C-banding polymorphism in one population of Aegilops comosa subsp. comosa var. comosa (Poaceae). Plant Syst Evol 180:105–114
Gill BS, Sharma HC, Raupp WJ, Browder LE, Hatchett JH, Harvey TL, Moseman JG, Waines JG (1985) Evaluation of Aegilops species for resistance to wheat powdery mildew, wheat leaf rust, Hessian fly, and greenbug. Plant Dis 69:314–316
Gong WP, Li GR, Zhou JP, Li GY, Liu C, Huang CY, Zhao ZD, Yang ZJ (2014) Cytogenetic and molecular markers for detecting Aegilops uniaristata chromosomes in a wheat background. Genome 57:481–489
Hao M, Luo JT, Yang M, Zhang LQ, Yan ZH, Yuan ZW, Zheng YL, Zhang HG, Liu DC (2011) Comparison of homoeologous chromosome pairing between hybrids of wheat genotypes Chinese Spring ph1b and Kaixian-luohanmai with rye. Genome 54:959–964
Huang XQ, Cloutier S, Lycar L, Radovanovic N, Humphreys DG, Noll JS, Somers DJ, Brown PD (2006) Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor Appl Genet 113:753–766
Huang L, He Y, Jin YR, Wang F, He JS, Feng LH, Liu DC, Wu BH (2018) Characterization of novel LMW glutenin subunit genes at the Glu-M3 locus from Aegilops comosa. 3. Biotech 8:379
Iqbal N, Reader SM, Caligari PD, Miller TE (2000) Characterization of Aegilops uniaristata chromosomes by comparative DNA marker analysis and repetitive DNA sequence in situ hybridization. Theor Appl Genet 101:1173–1179
Ishikawa G, Nakamura T, Ashida T, Saito M, Nasuda S, Endo TR, Wu JZ, Matsumoto T (2009) Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor Appl Genet 118:499–514
Kawahara T (2000) Isozyme variation in species of the section Comopyrum of Aegilops. Genet Resour Crop Ev 47:641–645
Kilian B, Mammen K, Millet E, Sharma R, Graner A, Salamini F, Hammer K, Özkan H (2011) Chapter 1. Aegilops. In: Kole C (ed) Wild crop relatives: genomic and breeding resources. Cereals, Springer, New York, pp 1–76
Kimber G, Sears ER (1983) Assignment of genome symbols in the Triticeae. In: Sakamoto S (ed) Proceedings of the 6th international wheat genetics symposium. Kyoto, Japan, pp 1195–1196
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137
Kuraparthy V, Sood S, Chhuneja P, Dhaliwal HS, Kaur S, Bowden RL, Gill BS (2007) A cryptic wheat-Aegilops triuncialis translocation with leaf rust resistance gene Lr58. Crop Sci 47:1995–2003
Liu C, Gong WP, Han R, Guo J, Li GR, Li HS, Song JM, Liu AF, Cao XY, Zhai SN, Cheng DG, Li GY, Zhao ZD, Yang ZJ, Liu JJ, Reader SM (2019) Characterization, identification and evaluation of a set of wheat-Aegilops comosa chromosome lines. Sci Rep 9:4773
McIntosh RA, Miller TE, Chapman V (1982) Cytogenetical studies in wheat XII. Lr28 for resistance to Puccinia recondita and Sr34 for resistance to P. graminitritici. Z. Pflanzenzucht 89:295–306
Millet E, Rong JK, Qualset CO, Mcguire PE, Bernard M, Sourdille P, Feldman M (2014) Grain yield and grain protein percentage of common wheat lines with wild emmer chromosome-arm substitutions. Euphytica 195:69–81
Mohler V, Albrecht T, Castell A, Diethelm M, Schweizer G, Hartl L (2016) Considering causal genes in the genetic dissection of kernel traits in common wheat. J Appl Genet 57:467–476
Molnár I, Kubaláková M, Šimková H, Cseh A, Molnár-Láng M, Doležel J (2011) Chromosome isolation by flow sorting in Aegilops umbellulata and Ae. comosa and their allotetraploid hybrids Ae. biuncialisand Ae. geniculata. PLoS One 6:e27708
Molnár I, Šimková H, Leverington-Waite M, Goram R, Cseh A, Vrána J, Farkas A, Doležel J, Molnár-Láng M, Griffiths S (2013) Syntenic relationships between the U and M genomes of Aegilops, wheat and the model species brachypodium and rice as revealed by COS markers. PLoS One 8:e70844
Rakszegi M, Molnár I, Lovegrove A, Darkó É, Farkas A, Láng L, Bedő Z, Doležel J, Molnár-Láng M, Shewry P (2017) Addition of Aegilops U and M chromosomes affects protein and dietary fiber content of wholemeal wheat flour. Front Plant Sci 8:1529
Riley R, Chapman V, Johnson R (1968) Introduction of yellow rust resistance of Aegilops comosa into wheat by genetically induced homoeologous recombination. Nature 217:383–384
Schneider A, Linc G, Molnár I, Molnár-Láng M (2005) Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat-Aegilops biuncialis disomic addition lines. Genome 48:1070–1082
Sharma SN, Sain RS, Sharma RK (2003) Genetics of spike length in durum wheat. Euphytica 130:155–161
Song ZP, Dai SF, Bao TY, Zuo YY, Xiang Q, Li J, Liu G, Yan ZH (2020) Analysis of structural genomic diversity in Aegilops umbellulata, Ae. markgrafii, Ae. comosa, and Ae. uniaristata by fluorescence in situ hybridization karyotyping. Front. Plant Sci 11:710
Tang ZX, Yang ZJ, Fu SL (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318
Tanguy AM, Coriton O, Abélard P, Dedryver F, Jahier J (2005) Structure of Aegilops ventricosa chromosome 6Nv, the donor of wheat genes Yr17, Lr37, Sr38, and Cre5. Genome 48:541–546
Van Slageren MW (1994) Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. &Spach) Eig (Poaceae). Wageningen: Agricultural University Papers, the Netherlands, International Center for Agricultural Research in Dry Areas, Aleppo, Syria pp. 126–128
Wang K, Gao LY, Wang SL, Zhang YZ, Li XH, Zhang MY, Xie ZZ, Yan YM, Belgard M, Ma WJ (2006) Phylogenetic relationship of a new class of LMW-GS genes in the M genome of Aegilops comosa. Theor Appl Genet 122:1411–1425
Yan ZH, Wan YF, Liu KF, Zheng YL, Wang DW (2002) Identification of a novel HMW glutenin subunit and comparison of its amino acid sequence with those of homologous subunits. Chin Sci Bull 47:220–225
Yen C, Yang JL (2020) Biosystematics of Triticeae Vol. 1, Triticum-Aegilops complex. China Agricultural Press, Springer, pp 96–102 146–149
Zhao JX, Wang XJ, Pang YH, Cheng XN, Wang LM, Wu J, Yang QH, Chen XH (2016) Molecular cytogenetic and morphological identification of a wheat-L.mollis1Ns(1D) substitution line, DM45. Plant Mol Biol Report 34:1146–1152
Zhou JP, Cheng Y, Zang LL, Yang EN, Liu C, Zheng XL, Deng KJ, Zhu YQ, Zhang Y (2016) Characterization of a new wheat-1M(1B) substitution line with good quality-associated HMW glutenin subunit. Cereal Res Commun 44:198–205
Zhukovsky PM (1928) A critical-systematical survey of the species of the genus Aegilops L. Bull Appl Bot Genet Plant Breed 18:417–609
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This study is supported by the Sichuan Science and Technology Program (No. 2018HH0130 and No. 2018HH0113), the Natural National Science Foundation of China (31771783, U1403185), the Ministry of Science and Technology of China (2016YFD0100502, 2017YFD0100903), and the Key Research and Development Program of Sichuan Province (2018NZDZX0002).
Author information
Authors and Affiliations
Contributions
KL Zhu wrote the paper. KL Zhu, SF Dai, ZP Song, Q Xiang, YY Zuo, and TY Bao conducted experiments and data analysis. Gang L and Jian L conduct field experiment. GY Chen, YM Wei, and YL Zhang give some suggestion. ZH Yan conceived and designed the study.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kang, L., Dai, S., Song, Z. et al. Production and characterization of a disomic 1M/1D Triticum aestivum-Aegilops comosa substitution line. Mol Breeding 41, 16 (2021). https://doi.org/10.1007/s11032-021-01207-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01207-2