Abstract
Durum wheat (Triticum durum Desf.) is a major crop of North Africa. Here, its production is affected by Hessian fly (Mayetiola destructor, HF) epidemics. Genetic resistance against this pest exist, but its molecular basis remains unclear. Here, a panel of 159 modern durum lines were exposed to the Moroccan HF biotype. Association mapping studies revealed three major loci conferring resistance. QH.icd-2A was identified at LOD of 24.1 on the telomeric end of 2AL, and it is believed to represent a novel locus derived from T. dicoccum. QH.icd-5B was identified on 5BS at LOD of 9.5, and it appears as overlapping with H31. QHara.icd-6B was identified at LOD of 54.5. This locus confers resistance to five Moroccan released cultivars, including ‘Faraj’. A mapping population (MP) was obtained by crossing ‘Faraj’ and a HF-susceptible cultivar ‘Gidara2’. Challenging this MP with the Moroccan and Great Plains HF biotypes revealed 1:1 segregation ratio fitting of a single gene. Quantitative trait locus (QTL) analysis confirmed a single locus at LOD of 35 and 25 for the two biotypes, respectively, corresponding to QHara.icd-6B. This locus spans a 7.7 cM interval, and it is derived via introgression from a resistant T. araraticum. One KASP marker (BS00072387) was validated for use in breeding on a separate set of elite lines, to show r2 of 0.65 and accuracy of 0.98. Finally, field testing across sites did not identify any yield drag for QHara.icd-6B. The work presented here provides ideal tools to incorporate HF-resistant loci in durum cultivars via marker-assisted breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Durum wheat (Triticum turgidum L. ssp. durum) is one of the main sources of calories and proteins in many developing countries, especially in Mediterranean semi-arid areas (Magallanes-López et al. 2017; Bassi and Sanchez-Garcia 2017). However, this crop is affected by several abiotic and biotic stresses. Among these, Hessian fly (HF), Mayetiola destructor (Say), is the most damaging insect pest, particularly in North Africa, North of Kazakhstan, South Europe and North America (El Bouhssini et al. 2012). In Morocco, this insect causes 32–36% wheat yield losses during dry years (Lhaloui et al. 1992; Nsarellah et al. 2003; Jlibene and Nsarellah 2011). In the USA, losses caused by Hessian fly have been estimated at $100 million per year (Berzonsky et al. 2003). Several integrated pest management strategies have been tried to manage HF populations combining anticipated planting date, crop rotation, sex pheromone traps, plowing or burning of stubble, and the use of systemic insecticides. However, the most effective approach remains host plant resistance, which is the most practical, environmentally sound, and economical option for HF control (Ratcliffe and Hatchett 1997; Berzonsky et al. 2003; Harris et al. 2015).
A gene-for-gene relationship was demonstrated between resistance in wheat and virulence in Hessian fly. Resistance in wheat is conditioned mostly by dominant alleles at one or a pair of loci, with the exception of h4 that is recessive in nature. The virulence against each resistance wheat allele is conditioned by recessive alleles at a single locus in the Hessian fly genome (Hatchett and Gallun 1970; Zhao et al. 2016).
To date, 35 HF resistance genes have been identified in wheat and its wild relatives (Li et al. 2015; Tang et al. 2018). H1, H2, H3, h4, H5, H7, H8, H12, H32, and H34 were derived from common wheat; H13, H22, H23, H24, and H26 were introgressed from Aegilops tauschii; H6, H9, H10, H11, H14, H15, H16, H17, H18, H19, H20, H28, H29, and H31 originated in durum wheat; H21 and H25 are translocations from rye (Secale cereale), while H27, H30, and Hdic originated in Ae. ventricosa, Ae.triuncialis, and a cultivated primitive emmer wheat (Triticum turgidum ssp. dicoccum), respectively (Liu et al. 2005; Sardesai et al. 2005; Li et al. 2013). These resistance genes were mapped to various wheat chromosomes using molecular markers. The first gene mapped was H6 on chromosome 5A through monosomic analysis (Gallun and Patterson 1977). H3, H5, H6, H9, H10, H11, H12, H13, H14, H16, and H17 were mapped to chromosome 1AS using randomly amplified polymorphic DNA (RAPD) analysis (Wang et al. 2006). The resistance gene Hdic was also localized on chromosome 1AS using simple sequence repeat (SSR) markers (Liu et al. 2005). Restriction fragment length polymorphism (RFLP) markers are linked to H23, H24, and H25 genes on chromosome 6D, 3DL, and 6RL, respectively (Ma et al. 1993; Delaney et al. 1995). Seo et al. (1997) identified RAPD markers that are linked to the gene H21. The resistance gene H31 was mapped to the short arm of chromosome 5B with AFLP and STS markers (Williams et al. 2003). Sardesai et al. (2005) identified SSR markers that are associated with H32 on chromosome 3D. The resistance gene H34 was mapped to chromosome 6B using single nucleotide polymorphism markers (SNP) (Li et al. 2013).
Out of the 35 HF resistance genes identified so far, only H5, H11, H13, H14, H15, H21, H22, H23, H25, H26, and H34 are effective against the Moroccan fly populations (El Bouhssini et al. 2009). The results of screening wild relatives of wheat showed large number of resistant accessions of Aegilops tauschii and very limited sources of resistance in wild Triticum (El Bouhssini et al. 1998, 2008). However, screening of Triticum araraticum accessions at Kansas State University allowed the identification of several resistance sources (Gill et al. 1983). Four of these T. araraticum resistant sources were brought from the USA to Morocco, and crossed to locally adapted elites. Ten years of breeding selection resulted in 2003 in the inscription on the Moroccan catalog of four varieties of durum wheat cultivars resistant to HF: ‘Irden’, ‘Nassira’, ‘Marouane’, and ‘Chaoui’ (Nsarellah et al. 2003; Jlibene and Nsarellah 2011). Still, most of the released lines suffered from susceptibility to the fungal disease leaf rust (Puccinia triticina) and their yields were moderate. The collaboration between ICARDA and INRA resulted in 2007 in one additional release for Morocco: ‘Faraj’, a cultivar derived from the ‘Irden’ lineage, crossed to ICARDA elite lines, which combines resistant to HF with the capacity to withstanding leaf rust epidemics (Jlibene and Nsarellah 2011).
The deployment of wheat cultivars with high level of antibiosis to first instars exerts a strong selection pressure on Hessian fly populations, which favors selection of biotypes capable of surviving on resistant wheat. Several biotypes that vary in their ability to survive on wheat containing different genes have been identified (Gallun 1977; Ratchliffe et al. 2000; Naber et al. 2003; El Bouhssini et al. 2009). The Syrian HF population seems to be the most virulent as only two genes (H25 and H26) are effective (El Bouhssini et al. 2009). To stay ahead of HF biotype evolution, searching for new genes of resistance needs to be a continued effort. Hence, the objective of this study was to determine which resistant genes are currently deployed by Mediterranean breeders within a large international panel of durum wheat germplasm, and to specifically dissect the genomic region for resistance derived from T. araraticum.
Material and methods
Plant material
Three different sets of durum wheat germplasm were included in this study: a genome-wide association (GWAS) mapping panel, a bi-parental mapping population, and a set of ICARDA elite lines for validation. A total of 159 modern cultivars and elite lines were selected among 384 entries described in Kabbaj et al. (2017) and Sall et al. (2018a). The full panel included 10 sub-groups identified on the basis of genetic similarities (Kabbaj et al. 2017), of which four were composed of landraces and six of modern lines from ICARDA, CIMMYT, and eight other breeding programs. Since the scope of this study was specifically to dissect the source of resistance derived from T. araraticum, only 159 modern entries were used here. These belonged to the sub-group 7, which includes the Moroccan germplasm ‘Marouane’, ‘Amria’, and ‘Chaoui’ known for resistance to HF, and the sub-group 9, which includes the Moroccan cultivars ‘Nassira’ and ‘Faraj’-derived germplasm of ICARDA. Secondly, a mapping population was derived by crossing the HF-resistant cultivar ‘Faraj’ carrying a T. araraticum segment with ‘Gidara 2’, a HF susceptible cultivar released in Turkey with pedigree Stojocri/Omrabi3. A total of 150 F7 recombinant inbred lines (RIL) were obtained from this cross. Each individual plant was further multiplied to generate the 143 F7:12 plus the two parents that constitute the population under study. The third and final set was used for markers validation and it was composed of 94 ICARDA’s elites that constituted the 2017 international nurseries 40th IDYT and 40th IDON. Among these were 26 elites with ‘Faraj’ in their pedigree. In this article, the presence of ‘Faraj’ in the pedigree is presented by the breeders code ‘IcamorTA’, which was used before the actual release of the cultivar with this name. Similarly, the presence of the T. araraticum segment in the cultivar ‘Chaoui’ is indicated by the code ‘JSMor2BC1F1’, which refers to the original back-cross (BC) made with the T. araraticum lineage of ‘Irden’ identified in Jemaa Shaim station in Morocco (JSMor2).
The assessment of the dominance or recessive gene action of the main quantitative trait locus (QTL) identified here was assessed on four F2 populations obtained by crossing the resistant line ‘Icamoram7’ to four known susceptible parents: three landraces (ig:90264, 149_17, 175_13) and one Moroccan cultivar (Luiza).
Phenotyping
The Faraj/Giadara2 and GWAS panels were initially screened in the field at the experimental station of Jemaa Shaim (32°24.076; 008°46.976; 173 m) INRA-Morocco where HF epidemics occur regularly (Fig. 1). To coincide young stages of the crop with the second HF generation, the genotypes were late planted in early February 2015. The experiment was carried out using alpha lattice design with three replications. ‘Faraj’ and ‘Karim’ varieties were used as resistant and susceptible checks, respectively. Each accession was planted in 1-m row with 0.3-m inter-row spacing. Eight weeks later, the evaluation of the durum wheat population for resistance to HF was assessed, with susceptible plants showing clear stunting and contained live larvae.
Phenotypic effect of Hessian fly (HF) on durum wheat. a A close-up of the damaging pest Mayetiola destructor (Say). b The clear distinction between susceptible and resistant genotypes in seedling screening and c in field testing at Jemaa Shaim station with delayed planting. d Productivity changes between resistant and susceptible cultivars under normal planting with delayed rains. Same plot size and agronomic practices were used for the Moroccan main cultivar ‘Karim’ and the recent release ‘Faraj’
Additionally, all entries were planted under greenhouse controlled conditions at 23 ± 2 °C and a relative humidity of 65 ± 5%. The genotypes were sown in a standard plastic flat (54 × 36 × 8 cm) containing a mixture of 2/3 soil and 1/3 peat and divided into two longitudinal sections of 12 rows. Twenty seeds of each entry were sown in each row. In the middle of each flat, ‘Faraj’ and ‘Karim’ were used as resistant and susceptible checks, respectively. When the seedlings reached the one-leaf stage, the flats were covered with a cheesecloth tent, and about 50 mated females collected from the Chaouia region (Jemaa Shaim) were released and allowed to lay eggs for 3 days. Three weeks after the eggs hatched, the plant response to larval feeding was evaluated. Susceptible plants showed stunting and a dark green color and contained live larvae; whereas, the resistant plants exhibited a normal growth and a light green color and contained dead first-instar larvae (Fig. 1). Resistant plants were dissected under a binocular and checked for the presence of dead larvae to confirm the antibiosis effect. Only genotypes that presented resistant or susceptible response for all tested seedling and matched field resistance were declared as “resistant” or “susceptible”. The phenotyping procedure was repeated until all tested lines could be declared without possibility of error. This methodology of repeated phenotyping results in Hessian fly response scores of rather complete immunity (R-types) without any damage caused by the insect and dead larvae, or complete susceptibility (S-types), with all individuals showing stunted habitus with live larvae.
The Faraj/Gidara2 mapping population was also phenotyped under controlled conditions in the greenhouse in Manhattan, KS, using the Great Plains (GP) HF biotype (Ratchliffe et al. 2000). The screening test was carried out using the same procedure as above.
The Faraj/Gidara 2 mapping populations was yield tested in the field during seasons 2013–14 (14) and 2014–15 (15) at one site the first season and three sites the second season. The locations included three Moroccan sites prone to Hessian fly damage: Marchouch 14 and Marchouch 15 (33° 34′ 3.1″ N, 6° 38′ 0.1″ W, clay vertisol), and Sidi el Aydi 15 (33° 9′ 36″ N, 7° 24′ 0″ W, vertisol), and one Lebanese site were HF damage does not occur: Terbol (33° 48′ 29″ N, 35° 59′ 22″ W, chromic vertisol). The trials were planted following an augmented design with seven sub-blocks of size 24, each including four replicated checks. The plot size was 3.6 m2 at a sowing density of 120 Kg ha−1, of which 2.4 m2 were harvested, weighted, and converted to grain yield (kilogram per hectare). Agronomic practices varied based on the station but followed the general guidelines of timely sowing between the 15th of November and the 5th of December in Marchouch 14 and Terbol 15, while delayed sowing between the 10th of December and the 20th of December occurred in Marchouch 15 and Sidi el Aydi 15. A base pre-sowing application of 50 Kg ha−1 of N, P, and K. At stage 14 of Zadok’s scale herbicide was applied in a tank mixture to provide protection against both monocots and dicots. A week after herbicide application, ammonium nitrate was provided to add 36 Kg ha−1 of N. For Terbol 15 and Marchouch 14, a final application of urea was used to add 44 Kg ha−1 of N before flowering (Z65). All sites were rainfed, with the exception of Terbol 15, where one sprinkle irrigation of 20 mm was provided after flowering (Z65).
The F2 populations for evaluation of gene action were sown in growth chamber controlled conditions at 23 ± 2 °C and a relative humidity of 65 ± 5%. The genotypes were sown in a standard plastic flat (54 × 36 × 8 cm) containing a mixture of 2/3 soil and 1/3 peat. Twenty seeds of each entry were sown in each tray, for a total of three trays. In the middle of each flat, ‘Faraj’ and ‘Karim’ were used as resistant and susceptible checks, respectively. When the seedlings reached the one-leaf stage, the flats were covered with a cheesecloth tent, and about 50 mated females collected from the Chaouia region (Jemaa Shaim) were released and allowed to lay eggs for 3 days. Three weeks after the eggs hatched, the plant response to larval feeding was evaluated. Susceptible plants showed stunting and a dark green color and contained live larvae, whereas the resistant plants exhibited a normal growth and a light green color and contained dead first-instar larvae. Resistant plants were dissected under a binocular and checked for the presence of dead larvae to confirm the antibiosis effect. Not all sown seeds germinated properly, and only vigorous plants were included in the screening. The three replications were scored independently, and the amount of resistant and susceptible progenies of each cross was measured.
Genotyping
The association mapping panel was genotyped using the Axiom 35K “Breeders array” at TraitGenetics (Germany) following the provider guidelines. Details of this genotyping step have been previously discussed in Kabbaj et al. (2017) and Sall et al. (2018a). Briefly, 7652 high-fidelity polymorphic SNPs were obtained, with less than 1% missing data, and a minor allele frequency superior to 0.5% (i.e., at least two entries). The sequences of these markers were aligned with a cut-off of 98% identity to the International Wheat Genome Sequencing Consortium RefSeq ‘Chinese Spring’ genome assembly version 1.0 (IWGSC 2014; available at: https://wheat-urgi.versailles.inra.fr), to reveal their physical position. A sub-set of 500 highly polymorphic SNPs were selected on the basis of even spread along the genome, and used to identify the existence of population sub-structure, which reveal the existence of 10 main sub-groups (Kabbaj et al. 2017). To avoid bias, these 500 markers were then removed from all downstream analysis. Linkage disequilibrium was calculated as squared allele frequency correlations (r2) in TASSEL V 5.0 software (Bradbury et al. 2007), using the Mb position of the markers along the bread wheat reference genome. Linkage disequilibrium (LD) decay was estimated and plotted using the “Neanderthal” method (Jujumaan 2017). The LD decay was measured at 51.3 Mb for r2 < 0.2 (Fig. S1).
F7 seeds of the Faraj/Gidara 2 mapping population were grown under greenhouse conditions at the University of Jordan. Fresh leaves were collected and used for CTAB DNA extraction, and the DNA was then provided to TraitGenetics (Germany) for genotyping with a proprietary 20K SNP array. This array includes the most polymorphic Illumina 90K and Axiom 35K sequences.
For markers underlining QTLs of interest, the Axiom 35K or 20K array marker sequences were retrieved. The positive allele was identified for each marker by investigating the score of the resistant lines carrying ‘Faraj’ (coded as ‘IcamorTA’, which is a breeders’ code that stands for “ICARDA Morocco Triticum araraticum”) in the pedigree. These sequences were provided to LGC Genomics (UK), to design KASP markers using allele specific primers using the Primer3 software. Primer sequences of the suitable markers are presented in Table S1. These primers were then used to amplify the validation set following LGC standard procedures.
Data analysis
Seedling screening for HF relied only on scores that showed consistent results (resistant or susceptible) across all the 20 tested seedlings. Any genotype for which the score deviated from the others was repeated. Hence, perfect fit statistics was deemed unnecessary for the HF seedling screening. For field performances of the mapping population, best linear unbiased estimators (BLUEs) were obtained for each individual environment using the DAU.test option of the agricolae package (De Mendiburu 2016) on RStudio v1.0.136. Significance of the resistant vs. susceptible classes was obtained by grouping genotypes on the basis of their HF response in seedling, and considering the BLUE of the genotypes as replicates. The grain yield performances were then investigated among classes within environment by ANOVA using the lm and anova options of RStudio v1.0.136. Since significance at 0.01 level was identified for the classes, the F-protected option LSD test of the package agricolae (De Mendiburu 2016) on RStudio v1.0.136 was used to determine actual LSD differences between classes in each environment.
Genome-wide association analysis was conducted for the HF seedling score among the set of 159 modern lines, using the kinship matrix developed by Kabbaj et al. (2017) for this panel, but including only two sub-groups (clusters 7 and 9). The PROC-GLM function including the kinship matrix indicated above (Q + K) was used on the software TASSEL (Bradbury et al. 2007) to identify marker-traits associations. The value calculated for the LD decay of 51.3 Mb (Fig. S1) indicated that this association panel interrogated the 12,000 Mb of the durum wheat genome via 248 marker-traits hypothesis, and hence this panel has a Boneforroni correction as suggested by Duggal et al. (2008) for p < 0.01 equal to 3.4 LOD. In addition, Pearson’s critical values (Pearson 1985) for correlations r was squared to obtain a critical r2 of 0.053, and used to determine significant effect on phenotypic variation. Any marker-traits associations (MTAs) with LOD and r2 superior to these values was considered valid and presented here.
The genetic map of the Faraj/Gidara 2 population was generated as follows. First, all markers were aligned to the International Wheat Genome Sequencing Consortium RefSeq ‘Chinese Spring’ genome assembly version 1.0 (IWGSC 2014; available at: https://wheat-urgi.versailles.inra.fr), to reveal their physical position, using identity of 98% as cut-off value to remove wrong alignments. The correctly aligned markers were then sorted following the genomic order, and the options mrkdouble, mrkmerges, and squeeze of the software Carthagene (De Givry et al. 2005) at LOD of 3.0 were used to remove any marker that did not fit the order. Then flips with a window size of 9 and LOD of 3.0 was used to adjust the short distance errors in marker order. Thus, a map containing 1267 markers assigned to the 14 durum chromosomes was considered as the anchor on the software QTL IcIMapping v4.1 (Wang et al. 2016). The remaining markers were grouped at LOD of 5.0 and mapped using the By Anchor Order option. The resulting genetic map is used here even though, due to physical anchoring, the centromeric recombination distortions were not accounted for (see result section for details). The associations between markers and two phenotypes were analyzed in the same software using SMA option for single marker analysis and ICIM option for composite interval mapping with a step size of 0.5 cM. The Boneferroni correction (Duggal et al. 2008) was applied considering the 685 unique loci of the genetic map, to set a false discovery rate threshold at LOD of 3.8 for p < 0.01. In addition, Pearson’s table of critical values (Pearson 1985) was used to determine a cut-off for p < 0.01 equal to r2 = 0.055 as the minimal explanation of phenotypic variation by any QTLs.
The primers designed at LGC were used to genotype the validation set. For each marker that amplified and showed polymorphism, the regression cut-off was imposed at r2 = 0.105 following Pearson’s critical value (Pearson 1985) for association with the phenotype. In addition, accuracy was calculated as the ratio of the correct allelic call among all, sensitivity as the ratio of the correct positive allelic calls among all, and specificity as the ratio of the correct negative allelic calls among all.
The dominant or recessive nature of the main QTL identified here was assessed by χ2 test, considering as the expected frequency 0.75 and 0.25 of resistant plant in the case of dominant or recessive gene action, respectively. Each replication for each population was tested separately, also the average of each population and overall value were tested at p < 0.01.
Results
Phenotypic variation in resistance to Hessian fly and effect on yield performances
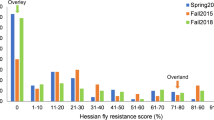
A total of 159 modern elites derived from eight breeding programs were assessed for response to a Moroccan Hessian fly population in wheat seedlings (Table 1). Among the six entries that were resistant to Hessian fly, five could trace their resistance to the T. araraticum introgression, including the Moroccan cultivars ‘Chaoui’, ‘Nassira’, and ‘Marouane’, and the elite lines ‘Icamoram7’ and ‘DWHF02’, which were obtained by crossing with ‘Faraj’. The final resistant elite line was ‘DWHF01’, a top cross derived from T. dicoccum (Sall et al. 2018b; Zaïm et al. 2017). The source of HF resistance in DWHF01is unknown. Among the susceptible group, 11 lines were derived from crosses with ‘Faraj’ but did not inherit the resistance to HF. Also, the cultivar ‘Amria’, supposedly carrying H5, exhibited a susceptible phenotype to the Moroccan HF biotype used here.
The mapping population Faraj/Gidara2 was also screened against both Moroccan and American (GP biotype) HF populations. A total of 52.4% and 38.7% resistant lines were identified as resistant against the two fly populations, respectively. A total of 16 lines exhibited different response between the two biotypes. The Mendelian segregation was analyzed by χ2 to reveal a 1:1 segregation (p < 0.01) in both cases, indicating a typical single gene inheritance not undergoing any selection bias. The same mapping population was also tested for yield performances at four different sites, three of which were mildly affected by HF. The individuals were divided into classes based on the response to the Moroccan HF population. ANOVA revealed significant (p < 0.01) effects for the classes, and the environment effects Fig 2 show the yield performances of the two classes in each environment. Less than one LSD difference was identified between the means of the two classes in Marchouch 14 and Terbol 15, while yield superiority of the resistant class could be shown in Marchouch 15 and Sidi el Aydi 15.
Grain yield performances across four field stations of progenies of the Faraj/Gidara 2 populations carrying (R-resistant) or not (S-susceptible) the T. araraticum segment. The solid black lines indicate the average, the box traces the 2nd and 3rd quartile, the whiskers indicate the 1st and 4th quartile, and empty circles represents outliers. The letter on the top of the figure indicate identical means (a), or means with differences superior to one LSD (a and b)
The validation set was also tested at seedlings stage against the Moroccan HF population. Six lines were resistant, and all of them were derived from crosses with ‘Faraj’. As for the association panel, 21 elites were derived from crosses with ‘Faraj’, but none of them inherited HF resistance.
Genetic dissection of Hessian fly resistance via association mapping
Association analysis applied to the panel identified three major MTAs. The main locus presented a LOD of 54.5 and explained 83% of the variation for HF response in general, and 100% of the variation for the five entries known to carry the T. araraticum segment based on pedigree (Table 2). This locus is linked to one single marker (AX-95181449) located on the telomere of chromosome 6BS, at a physical location 9 Mb away from the end of the chromosome. The temporary code (as per guidelines of the Catalog of Gene Symbols for Wheat, McIntosh et al. 2014) assigned to this locus is QHara.icd-6B, to represent H for Hessian fly resistance, ara for the T. araraticum origin, icd stands for ICARDA, and 6B is the chromosomal assignment. To assess the gene action of this major QTL, four F2 populations were derived by crossing four susceptible lines to the resistant line ‘Icamoram7’ harboring only the positive allele for QHara.icd-6B. Three independent replicates for each population were conducted and all confirmed dominant gene action via χ2 analysis (Table 3). In total, 181 F2 progenies were tested to reveal 73% and 27% HF resistant and susceptible, respectively. A near perfect match to the expected 75% and 25% frequencies for dominant effect.
The second major locus also linked to a single marker (AX-94980851) with a LOD of 24.1. It explained 32% of the total phenotypic variation, and only 9% of the variation for the lines containing T. araraticum in the pedigree (Table 2), with its presence limited to ‘DWHF01’ and ‘Nassira’. It is located in the telomeric portion of chromosome 2AL at position 520 Mb, and it did not originate from a T. araraticum translocation; thus, it was coded QH.icd-2A.
The third MTA was found on chromosome 5BS with a LOD of 9.5 (Table 2). It elapsed nine markers in linkage disequilibrium, and it explained 16% of the overall variation and 19% of the total phenotypic variation for the lines showing T. araraticum in the pedigree, with only ‘DWHF02’ carrying the positive allele for this locus. The markers’ physical location spans the very tip of the telomere of 5BS, from position 0.2 to 50 Mb; hence, it was coded QH.icd-5B.
A genetic map to confirm QHara.icd-6B
A genetic map was developed using the Faraj/Gidara 2 mapping population (Table 4). A total of 2067 markers were mapped, to generate 15 clusters corresponding to the 14 chromosomes of durum wheat, and one unassigned group. Chromosome 4A had the least number of markers assigned, while 2B had the highest, 33 and 290, respectively. The final map contained 685 unique loci, spanned 3587 cM, with chromosome 4A being the smallest at 137.3 cM and 2B the largest at 472.2 cM. The mapping procedure utilized did not account for the recombination distortion occurring at the centromere. In fact, the employed anchoring procedure relied on the physical position of specific markers and hence gaps superior to the normal unlinked cut-off of 50 cM still exist. As a result, the smallest centromere was identified for chromosome 6B and it enveloped just 18.5 cM, but the largest on chromosome 3A reached 147.2 cM. Hence, the overall mapping distance is over-estimated by 1011.3 cM, which brings the corrected map size to 2575.7 cM. This adjusted map size matches the 2592 cM reported by Maccaferri et al. (2014) for the durum wheat consensus map.
QTL analysis was conducted for both the Moroccan and American (GP) HF populations via single marker regression (SMR) and composite interval mapping (CIM). All methods and phenotypes identified the same unique telomeric region on chromosome 6BS (Fig. 4). The strongest association was identified by the Moroccan HF population, with LOD of 37 and 25 and it explained 79% and 54% of the total phenotypic variation, for CIM and SMR, respectively. Weaker but significant associations were also identified using the phenotypic data of the GP HF biotype, with LOD of 6 and r2 of 28% for both CIM and SMR. The overlap between the two QTL algorithms and two phenotypes pin-points to a segment comprised between 2.5 and 10.2 cM, corresponding to the physical position of 2.1–9.4 Mb on the IWGSC assembly. Unfortunately, the Axiom marker AX-95181449 identified in association panel was not included on the 20K array, so it is not possible to determine if this marker would have been significant also in the mapping population. Still, the physical position of AX-95181449 on the IWGSC assembly overlaps with this QTL interval; hence, it was also coded QHara.icd-6B. On the basis of its physical position, this marker was located in the genetic map in Fig. 3.
QTL analysis for Hessian fly (HF) response obtained via single marker regression (SMR) and composite interval mapping (CIM) for two biotypes one of Moroccan origin and one from Kansas. LOD and r2 are presented. The Bonferroni correction cut-off is displayed for the LOD. The light green square indicates the region of chromosome 6B where all models and phenotypes overlapped. The darker green area indicates the estimated genetic position of QHara.icd-6B identified by GWAS analysis (marker AX-95181449 indicated in red). The bolded marker name BS00072387 is the one that was successfully converted into KASP
Selection of markers for QHara.icd-6B detection
A total of 14 marker sequences were attempted for conversion to KASP primer combinations. Among these, six failed to generate primers that met the minimum in silico quality cut-off and were discarded before wet-lab testing. Of the remaining eight, two generated monomorphic calls for the validation set and were also discarded. Hence, only the results for six primers combinations are presented in Fig. 4. Unfortunately, five of the KASP markers did not correlate with the phenotype above the Pearson’s cut-off, even though BA00462979 reached acceptable levels of sensitivity, accuracy, and specificity. Only BS00072387 met all validation requirements, with r2 of 0.65, sensitivity of 0.67, accuracy of 0.98, and specificity of 1.00. This primer combination (sequences available in Table S1) was designed on the homonym marker at position 8.9 cM of the genetic map and at 2.1 Mb of the IWGSC genome, within the peak of QHara.icd-6B.
Validation on a set of 94 durum wheat elites of six KASP markers to follow the inheritance of QHara.icd-6B.The actual KASP score graph of the validation set is presented for BS00072387 as the haplotype distribution along the x- and y-axis of the HF resistant (solid black circle), HF susceptible (gray circle), and blank DNA (empty circle) as obtained from LGC SNP Viewer.
Discussion
Germplasm lines resistant to Hessian fly
Screening for HF resistant of a collection of 159 modern elite lines confirmed that resistance to this pest is rare, and only very targeted breeding efforts can deliver adapted and resistant cultivars. This is the case for the efforts conducted by the Moroccan durum wheat breeder, with the release of the cultivars ‘Faraj’, ‘Chaoui’, ‘Nassira’, and ‘Marouane’ (Jlibene and Nsarellah 2011), which confirmed their resistance in the screening conducted here. Study of the pedigree histories of these cultivars (Table 2) shows similarities only between ‘Chaoui’ and ‘Faraj’, both derived from crossing to a segregating progeny obtained from T. araraticum backcrossing, and tested at the Moroccan station of Jemaa Shaim, where HF epidemics are very common. The pedigree of ‘Nassira’ does provide evidence to its T. araraticum origin, and the creator of this line declared that it was among the funders of ‘Faraj’ (Jlibene and Nsarellah 2011). Further, the genetic study by Kabbaj et al. (2017) included ‘Nassira’ in sub-population 9, together with the ‘Faraj’-derived lines, while it placed ‘Marouane’ in sub-population 7. Similarly, Henkrar et al. (2016) confirmed strong genetic similarities between ‘Nassira’ and ‘Faraj’. ‘Marouane’ is the only one with no support in the pedigree for the T. araraticum source of resistance, nor similarity by genetic studies (Henkrar et al. 2016; Kabbaj et al. 2017). Yet, Jlibene and Nsarellah (2011) indicated that all Moroccan cultivars released in 2003, including ‘Marouane’, were derived from the same T. araraticum source, with the exception of ‘Amria’, which was derived from cross with a bread wheat containing H5. However, H5 is considered to be effective against the Moroccan biotypes, but ‘Amria’ was identified as susceptible against the Hessian fly population used in this study. Further investigation of the original seed set of ‘Amria’ revealed that this cultivar is in fact derived from the bulking of two genotypes: a HF-resistant and a HF-susceptible type (data not shown). As such, it can be concluded that the type used in this experiment was the susceptible one.
In addition, a previously uncharacterized resistant line (DWHF01) emerged from this study. This is a top cross between the elites ‘Younes’ and ‘Korifla’, both known to be susceptible, and an accession of T. dicoccum collected around Aleppo, Syria. Hence, it is possible that, as in the case of Hdic (Liu et al. 2005), this source of resistance was also originated from T. dicoccum. However, it is worth mentioning that Hdic itself does not provide resistance against the Moroccan HF biotype (El Bouhssini et al. 2009).
Localization of the novel Hessian fly resistance locus QH.icd-2A
QH.icd-2A is a major gene located at the telomere of 2AL controlling HF response in ‘DWHF01’ and possibly in ‘Nassira’. It does not explain significant variation for the T. araraticum source of resistance, but it otherwise explains 32% of the total phenotypic variation. On the basis of ‘DWHF01’ pedigree, it appears to have originated from a T. dicoccum accession collected from Syria. This locus does not appear to correspond to the previously characterized Hdic, which also originated from T. dicoccum, because Hdic was mapped to a different chromosome (1AS) and it does not provide resistance to the Moroccan biotype (Bouhssini et al. 2009). To the best of our knowledge, the only other HF gene reported thus far on chromosome 2A is the minor QTL QHf.osu-2A, identified in a mapping population of bread wheat (2174/Jagger) screened with biotype GP, and located at the telomere of chromosome 2AS (Tan et al. 2013). However, the QTL identified here and the one previously reported do not appear to overlap in terms of genomic location with QH.icd-2A located on 2AL, and QHf.osu-2A on 2AS. In addition, the resistance source also appears different, one derived from T. dicoccum from Syria and the second from a bread wheat cultivar. These considerations suggest that QH.icd-2A is indeed a novel HF-resistant gene, and that marker AX-94980851 can be used for its marker assisted selection starting from the elite line ‘DWHF01’.
Co-genetic localization of the Hessian fly resistance locus QH.icd-5B and H31
QH.icd-5B is a minor QTL located on the telomeric end of chromosome 5BS. It is underlined by a set of nine markers spanning 50 Mb and in strong linkage disequilibrium. Only the elite DWHF02 carries the positive allele at this locus. The pedigree of this ICARDA breeding line is extremely complex (Table 2) as it includes two primitive introgressions from T. araraticum and from T. polonicum L. from Turkey, together with several older elites. The main source of resistance carried by this line appears to be the T. araraticum segment, but it cannot be excluded that a second locus also contributes. In that sense, H31 was also identified on the very end of the short arm of chromosome 5B of a bread wheat cultivar, but its origin was in fact a Tunisian durum wheat landrace (Williams et al. 2003). As such, it is likely that QH.icd-5B corresponds to H31, and that this gene was introgressed to DWHF02 from the North African landraces background that constitute ‘Massara1’ (Mra1), ‘AinArous1’ (Aus1), or ‘Tensift1’. However, H31 is not effective against the Moroccan biotype, so even if these two loci are in fact the same gene, QH.icd-5B should represent a different allele. Alternatively, H31 in this background provides a corroborative role to the more effective QHara.icd-6B. In both cases, this locus appears of interest and more detailed co-segregation, and molecular study between ‘DWHF02’ and the original H31 donor Tunisian accession would be needed to clarify its role.
Detailed genetic localization of the Hessian fly resistance locus QHara.icd-6B
QHara.icd-6B was located as a major QTL with LOD of 54.5 at 9 Mb from the telomeric end of chromosome 6BS, and its dominant gene action was confirmed via F2 progeny testing (Table 3). It explained 100% of the phenotypic variation for the three Moroccan cultivars, ‘Chaoui’, ‘Marouane’, and ‘Nassira’, and the two ICARDA’s elites that inherited HF resistance from the Moroccan cultivar ‘Faraj’. These five entries also share only this locus as common source of resistance (Table 2) even though ‘Nassira’ also shares QH.icd-2A with ‘DWHF01’ and ‘DWHF02’ appears to also have a second resistant genes that overlaps with the position of H31. Since all five genotypes have inherited the HF resistance from a common T. araraticum source, but do not share any other evident pedigree history, it is considered that QHara.icd-6B maps in fact the original T. araraticum translocation.
To further characterize this key locus, a mapping population was generated between the resistant line ‘Faraj’ and the susceptible ‘Gidara 2’. Two biotypes were used to screen this RIL population and resulted in 75% agreement in response. The 25% differences identified are rather caused by minor experimental error due to small differences in procedures between the two laboratories, or by environmental variations as identified for other HF-resistant genes (Chen et al. 2014), or by true difference in the virulent spectrum of the two biotypes used. The last possibility seems unlikely since both biotypes identified one single segregating gene within the population by χ2 analysis, and QTL analysis confirmed it to be the exact same telomeric 6BS region of QHara.icd-6B identified via association mapping. This pin-pointed 7.7 cM interval corresponded to a span of 7.3 Mb on the IWGSC genome assembly and overlapped well with the physical position of 9 Mb of the marker AX-95181449. As expected, 79% and 28% of the phenotypic variation to the two HF biotypes were explained by this region, suggesting the presence of minor-genes effect that could not be identified via QTL analysis, or some minor degree of experimental error during the phenotyping phase. Nevertheless, extremely conservative statistical cut-off was applied to confirm that this region is indeed the translocation from T. araraticum that guarantees HF resistance to the Moroccan cultivars ‘Chaoui’, ‘Faraj’, ‘Marouane’, and ‘Nassira’.
T. araraticum has been identified in the past as an ideal source of resistance to HF, with 65 out of 147 accessions showed complete resistance (Gill et al. 1983). However, to the best of our knowledge, there is no other reported use of T. araraticum to provide HF resistance, other than the Moroccan released cultivars. In chromosome 6B, two other resistance genes H34 () and H25 are also assigned to this chromosome (Friebe et al. 1991; Li et al. 2013). H25 is a highly effective gene (Shukle et al. 2016) and was introgressed into bread wheat form the rye cultivar ‘Balbo’, via a terminal translocation T6BS·6BL-6R#2L. As such, the position of this gene corresponds to the telomeric end of 6BS where QHara.icd-6B localized. However, further characterization of this translocation located H25 to the distal end of 6RL (Delaney et al. 1995), which does not appear as the correct co-linear location on 6GS of T. araraticum of QHara.icd-6B. Also, while there could be some geographical overlap between rye and T. araraticum cultivation in Turkey, it would be hard to explain how the same resistant gene originated both on the R and G genomes, which share very limited ancestry. Finally, the only reported transfer of this 6RL segment in durum wheat cultivar ‘Cando’ utilized a 4AL translocation (Ti4AS·4AL-6R#1L-4AL; Friebe et al. 1999), rather than the 6BS one described above, and there is no evidence of parentage in the pedigree of ‘Faraj’. Hence, while more detailed studies would be required for confirmation, it is unlikely that the resistant gene presented here is in fact the same locus as H25. The second resistant gene located on 6B is H34, derived from the bread wheat variety ‘Clark’ (Foster et al. 1988). This gene was mapped in proximity of the centromere of chromosome 6B (Li et al. 2013) in a bread wheat RIL population, and the position of the SRR surrounding the peak was also assigned to the 6B centromere in the durum wheat consensus genetic map of Maccaferri et al. (2014). Further, Shukle et al. (2016) determined that this gene is not effective against the virulent HF biotypes used in this study. These observations, together with its origin from bread wheat rather than T. araraticum, do not support the hypothesis that QHara.icd-6B corresponds to H34. In absence of other reported HF genes in the literature, it appears that QHara-icd-6B is indeed a novel HF resistance gene.
Linkage drags and mode of inheritance of the QHara.icd-6B segment
The G genome of the Armenian wheat can pair to the B genome of durum, even though sterility often occurs (Brown-Guedira et al. 1997; Kilian et al. 2007). It is then unsurprising that this fragment was located at the telomere of 6BS, since telomeric region appears as more prone to recombine between G and B, as it was hypothesized for the introgression of Sr40 from T. araraticum to the telomere of 2BS of bread wheat (Dyck 1992).
More importantly, when attempting to breed an alien introgression, it is first necessary to understand the existence of possible linkage drags. To assess possible issues of yield drag, the mapping population was divided into classes based on the phenotypic response to Moroccan HF population and yield tested in four environments. At Marchouch 2014 and Terbol 2015, HF infestations were not evident due to good distribution of rainfall. Under these conditions, no yield disadvantage could be observed between lines with and without the resistance, suggesting no evident yield drag for the deployment of this QTL. Instead, at Marchouch 2015 and Sidi el Aydi 2015 where early-season droughts occurred, as shown by the lower yields, HF damages could be observed. Under these conditions, the resistant lines resulted in an average yield advantage of 21% and 11% for the two stations, respectively. Considerations for other traits were made by Taghouti et al. (2017) that concluded ‘Chaoui’, ‘Faraj’, ‘Marouane’, and ‘Nassira’ did represent an improvement for grain yield, earlier maturity, shorter stature, and similar gluten strength, but that they represented a reduction in the yellow color of the grains, an important quality trait for durum wheat. However, the same article reports that ‘Faraj’ had the highest score for yellow color among all released cultivars until 2007. As such, it is possible to conclude that the T. araraticum introgression does not cause any evident genetic drags for yield, phenology, or gluten strength, but that it might be associated with some negative effect on yellow pigment, a negative linkage that could be broken in the case of ‘Faraj’.
Concerning the inheritance of the trait, the mapping population showed normal Mendelian inheritance and the F2 progeny testing confirmed dominant gene effect. However, this study also showed that a total of 11 elites among the association panel and 21 elites in the validation set were derived from crosses to ‘Faraj’, but did not inherit the QHara.icd-6B segment. These represent a fourfold difference compared to the eight ‘Faraj’-derived elites that inherited the resistance. As such, it would appear that breeders’ selection not in the presence of the pest can cause rapid loss of this key trait. Conversely, it was noted that in many cases, the spike size of ‘Faraj’-derived lines tended to be smaller, but that this yield disadvantage was compensated by a higher number of tillers. If this was to be confirmed, then it could be hypothesized that during selection, breeders tend to discard progenies with smaller spikes and hence favor the loss of the HF resistance. More data are necessary to confirm this hypothesis, but these preliminary observations call for the development and deployment of easy-to-use markers for trait selection.
Validation of a KASP marker to select QHara.icd-6B
The genotyping platforms utilized here (Axiom 35K and 20K property array) have proven extremely effective in identifying the regions harboring QHara.icd-6B. However, their massive use in everyday marker assisted selection (MAS) breeding would be hampered by their high cost per sample. To provide a more scalable solution, 14 marker sequences within and surrounding the main QTL peak were used for conversion to KASP primers. Only one of these sequences (BS00072387) could be effectively converted and validated for MAS. This marker is located at position 8.9 cM of the mapping population, 1 cM away from the maximum QTL peak and well within the 7.7 cM interval of QHara.icd-6B. Yet, its physical position on the IWGSC genome assembly at 2.1 Mb is at the very telomeric end of the significant interval (2.1–9.4 Mb). Nevertheless, this marker predicted 65% of the phenotypic variation and it was 98% of the time accurate in calling the resistance mediated by T. araraticum in a validation set of elite lines completely different from the germplasm used for the association studies or the mapping population. Further, it did not cause any false positive with 100% specificity, but it did fail to recognize the resistance carried by two elite lines (67% sensitivity). Hence, it can be considered a solid and ready-to-use co-dominant marker to follow the inheritance of QHara.icd-6B.
Conclusion
In this study, we could identify via association mapping three major loci controlling HF resistance in durum wheat. Among these, one is likely to correspond to the already identified gene H31 on chromosome 5BS, while the two others appear to be novel: QH.icd-2A from T. dicoccum and QHara.icd-6B from T. araraticum. This latter locus is of great importance for ensuring HF resistance to four Moroccan cultivars, and as such was given greater priority. A mapping population was devised to confirm the results of the association mapping. In addition, no evident linkage drag was associated with the deployment of this 6G introgression, with the exception of a hypothesized change in spike size in favor of spike number. Further, a new KASP marker was validated to easily follow the inheritance of this trait via MAS. With these new tools in hand, breeders can now easily deploy these resistant genes via a targeted pyramiding scheme. However, it was also evidenced that with the exceptions of ‘DWHF02’, which likely carries also H31, and ‘Nassira’, which might integrate QH.icd-2A, all other Moroccan cultivars rely on a single HF-resistant gene. As Morocco prepares to roll out these varieties in large scale, it is important to rapidly pyramid additional resistant genes to prevent novel mutations in the pest that could overcome this precious source of resistance.
References
Bassi FM, Sanchez-Garcia M (2017) Adaptation and stability analysis of ICARDA durum wheat (Triticum durum Desf.) elites across 18 countries. Crop Sci 57:1–12. https://doi.org/10.2135/cropsci2016.11.0916.
Berzonsky WA, Ding H, Haley SD, Harris MO, Lamb RJ, McKenzie RIH, Ohm HW, Patterson FL, Peairs FB, Porter DR, Ratcliffe RH, Shanower TG (2003) Breeding wheat for resistance to insects. Plant Breed Rev 22:221–296
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brown-Guedira GL, Gill BS, Cox TS, Leath S (1997) Transfer of disease resistance genes from Triticum araraticum to common wheat. Plant Breed 116:105–112
Chen MS, Wheeler S, Wang H, Jeffrey R (2014) Impact of temperatures on hessian fly (Diptera: Cecidomyiidae) resistance in selected wheat cultivars (Poales: Poaceae) in the Great Plains region. J Econ Entomol 107:1261273. https://doi.org/10.1603/EC13357
De Givry S, Bouchez M, Chabrier P, Milan D, Schiex T (2005) CARTHAGENE: multipopulation integrated genetic and radiated hybrid mapping. Bioinformatics 21:1703–1704. https://doi.org/10.1093/bioinformatics/bti222
De Mendiburu F (2016) A statistical analysis tool for agricultural research. Dissertation, National Engineering University Lima-Peru
Delaney DE, Friebe BR, Hatchett JH, Gill BS, Hulbert SH (1995) Targeted mapping of rye chromatin in wheat by representational difference analysis. Genome 38:458–466
Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE (2008) Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 9:516
Dyck PL (1992) Transfer of a gene for stem rust resistance from Triticum araraticum to hexaploid wheat. Genome 35:788–792. https://doi.org/10.1139/g92-120
El Bouhssini M, Benlhabib O, Nachit M, Houari A, Bentika A, Nsarellah N, Lhaloui S (1998) Identification in Aegilops species of resistant sources to Hessian fly (Diptera: Cecidomyiidae) in Morocco. Genet Resour Crop Evol 45:343–345
El Bouhssini M, Nachit M, Valkoun J, Abdalla O, Rihawi F (2008) Sources of resistance to Hessian fly (Diptera: Cecidomyiidae) in Syria identified among Aegilops species and synthetic derived bread wheat lines. Genet Resour Crop Evol 55:1215–1219
El Bouhssini M, Chen M, Lhaloui S, Zharmukhamedova G, Rihawi F (2009) Virulence of Hessian fly (Diptera: Cecidomyiidae) in the Fertile Crescent. J Appl Entomol 133:381–385
El Bouhssini M, Ogbonnaya FC, Chen M, Lhaloui S, Rihawi F, Dabbous A (2012) Sources of resistance in primary synthetic hexaploid wheat (Triticum aestivum L.) to insect pests: Hessian fly, Russian wheat aphid and Sunn pest in the Fertile Crescent. Genet Resour Crop Evol 60:621–627
Foster JE, Ohm H, Shaner GE, Buechley G (1988) Registration of ‘Clark’ wheat. Crop Sci 28:1032. https://doi.org/10.2135/cropsci1988.0011183X002800060058x
Friebe B, Hatchett JH, Gill BS, Mukai Y, Sebesta EE (1991) Transfer of Hessian fly resistance from rye to wheat via radiation-induced terminal and intercalary chromosomal translocations. Theor Appl Genet 83:33–40
Friebe B, Kynast RG, Hatchett JH, Sears RG, Wilson DL, Gill BS (1999) Transfer of wheat-rye translocation chromosomes conferring resistance to Hessian fly from bread wheat into durum wheat. Crop Sci 39:1692–1696
Gallun RL (1977) Genetic basis of Hessian fly epidemics. Ann NY Acad Sci 287:223–229
Gallun RL, Patterson FL (1977) Monosomic analysis of wheat for resistance to Hessian fly. J Hered 68:223–236
Gill BS, Browder LE, Hatchett JH, Harvey TL, Raupp WJ, Sharma HC, Waines JG (1983) International Wheat Genetics Symposium. In: Proceedings of the sixth International Wheat Genetics Symposium. Maruzen Co., Kyoto
Harris MO, Friesen TL, Xu SS, Chen MS, Giron D, Stuart JJ (2015) Pivoting from Arabidopsis to wheat to understand how agricultural plants integrate responses to biotic stress. J Exp Bot 66:513–531
Hatchett JH, Gallun RL (1970) Genetics of the ability of the Hessian fly, Mayetiola destructor (say), to survive on wheats having different genes for resistance. Ann Entomol Soc Am 63:1400–1407
Henkrar F, El-Haddoury J, Ouabbou H, Nsarellah N, Iraqi D, Bendaou N, Udupa SM (2016) Genetic diversity reduction in improved durum wheat cultivars of Morocco as revealed by microsatellite markers. Sci Agric 73:134–141. https://doi.org/10.1590/0103-9016-2015-0054
IWGSC [International Wheat Genome Sequencing Consortium] (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
Jlibene M, Nsarellah N (2011) Wheat breeding in Morocco, a historical perspective. In: Angus W, Bonjean A, Van Ginkel M (eds) The World wheat book: a history of wheat breeding. Lavoisier, Paris, p. 425–442
Jujumaan (2017) Estimating and plotting the decay of linkage disequilibrium. https://jujumaan.com/2017/07/15/linkage-disequilibrium-decay-plot/. Accessed 15 July 2017.
Kabbaj SAT, Al-Abdallat A, Geleta M, Amri A, Filali-Maltouf A, Belkadi B, Ortiz R, Bassi FM (2017) Genetic diversity within a global panel of durum wheat (Triticum durum) landraces and modern germplasm reveals the history of alleles exchange. Front Plant Sci 18:1277. https://doi.org/10.3389/fpls.2017.01277
Kilian B, Özkan H, Deusch O, Effgen S, Brandolini A, Kohl J, Martin W, Salamini F (2007) Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol 24:217–227
Lhaloui S, Buschman L, El Bouhssini M, Amri A, Hatchett JH, Keith D, Starks K, El Houssaini K (1992) Control of Mayetiola species (Diptera: Cecidomyiidae) with carbofuran in bread wheat, durum wheat and barley, with yield loss assessment and its economic analysis. Awamia 77:55–73
Li C, Chen M, Chao S, Yu J, Bai G (2013) Identification of a novel gene, H34, in wheat using recombinant inbred lines and single nucleotide polymorphism markers. Theor Appl Genet 126:2065–2071
Li G, Wang Y, Chen MS, Edae E, Poland J, Akhunov E, Chao S, Bai G, Carver BF, Yan L (2015) Precisely mapping a major gene conferring resistance to hessian fly in bread wheat using genotyping-by-sequencing. BMC Genomics 16:108
Liu XM, Brown-Guedira GL, Hatchett JH, Owuoche JO, Chen MS (2005) Genetic characterization and molecular mapping of a hessian fly-resistance gene transferred from T. turgidum ssp. dicoccum to common wheat. Theor Appl Genet 111:1308–1315
Ma ZQ, Gill BS, Sorrells ME, Tanksley SD (1993) RFLP markers linked to hessian fly-resistance genes in wheat (Triticum aestivum L.) from Triticum tauschii (Coss.) Schmal. Theor Appl Genet 85:750–754
Maccaferri M, Cane’ MA, Sanguineti MC, Salvi S, Colalongo MC, Massi A, Clarke F, Knox R, Pozniak CJ, Clarke JM, Fahima T, Dubcovsky J, Xu S, Ammar K, Karsai I, Vida G, Tuberosa R (2014) A consensus framework map of durum wheat (Triticum durum Desf.) suitable for linkage disequilibrium analysis and genome-wide association mapping. BMC Genomics 15:873
Magallanes-López AM, Hernandez-Espinosa N, Velu G, Posadas-Romano G, McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xia XC (2017) Variability in iron, zinc and phytic acid content in a worldwide collection of commercial durum wheat cultivars and the effect of reduced irrigation on these traits. Food Chem 237:499–505. https://doi.org/10.1016/j.foodchem.2017.05.110
McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Xia XC (2014) Catalogue of gene symbols for wheat: 2013–14 supplement
Naber N, El Bouhssini M, Lhaloui S (2003) Biotypes of hessian fly (Diptera; Cecidomyiidae) in Morocco. J Appl Entomol 127:174–176
Nsarellah N, Amri A, Nachit M, El Bouhssini M, Lhaloui S (2003) New durum wheat with hessian fly resistance from Triticum araraticum and Triticum carthlicum in Morocco. Plant Breed 122:435–437
Pearson K (1985) Notes on regression and inheritance in the case of two parents. Proc R Soc Lond 58:240–242
Ratchliffe RH, Cambron SE, Flanders KL, Bosque-Perez NA, Clement SL, Ohm HW (2000) Biotype composition of Hession fly (Diptera: Cecidomyiidae) population from the southern, Midwestern, and north western United States and virulence to resistance genes in wheat. J Econ Entomol 93:1319–1328
Ratcliffe RH, Hatchett JH (1997) Biology and genetics of the Hessian fly and resistance in wheat’. In: Bondari K (ed) New Developments in Entomology. Research Signpost, Trivandum, pp 47–56
Sall AT, Bassi FM, Cisse M, Gueye H, Ndoye I, Filali-Maltouf A, Ortiz R (2018a) Durum wheat breeding: in the heat of the Senegal River. Agriculture 8:99. https://doi.org/10.3390/agriculture8070099
Sall AT, Cisse M, Gueye H, Kabbaj H, Ndoye I, Filali-Maltouf A, Belkadi B, El-Mourid M, Ortiz R, Bassi FM (2018b) Heat tolerance of durum wheat (Triticum durum Desf.) elite germplasm tested along the Senegal River. J Agric Sci 10:217. https://doi.org/10.5539/jas.v10n2p217
Sardesai N, Nemacheck JA, Subramanyam S, Williams CE (2005) Identification and mapping of H32, a new wheat gene conferring resistance to hessian fly. Theor Appl Genet 111:1167–1173
Seo YW, Johnson JW, Jarret RL (1997) A molecular marker associated with the H21 hessian fly resistance gene in wheat. Mol Breed 3:177–118
Shukle RH, Cambron SE, Moniem HA, Schemerhorn BJ, Redding J, Buntin GD, Flanders KL, Reisig DD, Mohammadi M (2016) Effectiveness of genes for hessian fly (Diptera: Cecidomyiidae) resistance in the southeastern United States. J Econ Entomol 109:399–405
Taghouti M, Nsarellah N, Gaboun F, Rochdi A (2017) Multi-environment assessment of the impact of genetic improvement on agronomic performance and on grain quality traits in Moroccan durum wheat varieties of 1949 to 2017. Global journal of plant breeding and. Genetics 4:394–404
Tan CT, Carver BF, Chen MS, Gu YQ, Yan L (2013) Genetic association of OPR genes with resistance to hessian fly in hexaploid wheat. BMC Genomics 14:369
Tang G, Liu X, Chen GH, Witworth RJ, Chen MS (2018) Increasing temperature reduces wheat resistance mediated by major resistance genes to Mayetiola destructor (Diptera: Cecidomyiidae). J Econ Entomol 111:1433–1438
Wang T, Xu SS, Harris MO, Hu J, Liu L, Cai X (2006) Genetic characterization and molecular mapping of hessian fly resistance genes derived from Aegilops tauschii in synthetic wheat. Theor Appl Genet 113:611–618
Wang J, Li H, Zhang L, Meng L (2016) Users’ manual of QTL IciMapping. The Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing 100081, China, and Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT), Mexico
Williams CE, Collier CC, Sardesai N, Ohm HW, Cambron SE (2003) Phenotypic assessment and mapped markers for H31, a new wheat gene conferring resistance to hessian fly (Diptera: Cecidomyiidae). Theor Appl Genet 107:1516–1523
Zaïm M, El Hassouni K, Gambac F, Filali-Maltouf A, Belkadi B, Sourourd A, Amri A, Nachit M, Taghouti M, Bassi FM (2017) Wide crosses of durum wheat (Triticum durum Desf.) reveal good disease resistance, yield stability, and industrial quality across Mediterranean sites. Field Crop Res 214:219–227
Zhao C, Shukle R, Navarro-Escalante L, Chen MS, Richards S, Stuart JJ (2016) Avirulence gene mapping in the hessian fly (Mayetiola destructor) reveals a protein phosphatase 2C effector gene family. J Insect Physiol 84:22–31
Acknowledgments
The authors wish to recognize the technical contribution by the durum wheat staff for field research. We wish also to thank Mme M. Zaim for the help provided with the association study.
Funding
This research was partially supported by funds from Australia’s Grain & Research Development Corporation (GRDC) grant “ICA00012: Focused improvement of ICARDA/Australian durum germplasm for abiotic tolerance” and CGIAR CRP wheat.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Table S1
(DOCX 14 kb)
Figure S1
Genome-wide average linkage disequilibrium (LD) decay over genetic distances. a) Plot of pair-wise LD r2 values as a function of inter-marker map distance (Mbp). The blue curve represents the model fit to LD decay. The red line represents the intercept to r2 = 0.2. b) Boxplot showing LD r2 values from pair-wise comparison of markers at increasing physical distances: 0–1, 2–51, 52–100, 101–200, 201–300, 301–400, 401–500 and > 500 Mbp. LD decay value was set at 51.3 Mb. (PNG 200 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Bassi, F.M., Brahmi, H., Sabraoui, A. et al. Genetic identification of loci for Hessian fly resistance in durum wheat. Mol Breeding 39, 24 (2019). https://doi.org/10.1007/s11032-019-0927-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-0927-1