Abstract
We identified haplotype-tagging insertion/deletions (InDels) for downy mildew resistance (Rpv3-1) in grapevine and converted them into InDel markers. InDel-25,941 and InDel-26,032 were validated by fragment analysis via capillary electrophoresis in 174 varieties of Vitis vinifera, 50 resistant varieties of the ‘Seibel 4614’ lineage that share Rpv3-1 by descent, and in 83 Vitis accessions. Amplicon sequencing of ancestral and derived alleles revealed that both mutations were caused by deletions. The 25,941-deletion is most likely recent. The derived allele is present only in resistant varieties obtained from ‘Seibel 4614’ and has originated in North American populations through two successive deletions within a predicted multiple stem-loop ssDNA structure, consisting of three nearby short inverted repeats, which shortened the ancestral DNA stepwise. The 26,032-deletion is more ancient. The derived allele is always present in resistant varieties of the ‘Seibel 4614’ lineage, completely absent from V. vinifera, not found in other North American accessions, and rarely present in Asian species. It may have originated in a common ancestral population before the continental disjunction, followed by incomplete lineage sorting, or in either lineage followed by introgression via secondary contacts. Genotyping with these markers does not require special instruments or chemistry for routine screening in breeding practice. Differences in amplicon size between grapes that carry or do not carry Rpv3-1 are detectable via standard agarose gel electrophoresis, or classical melting curve analysis using nonsaturating fluorescent dyes. The recombination rate between each marker and the trait locus is 0.118% for InDel-25,941 and 0.071% for InDel-26,032.




Similar content being viewed by others
References
Adam-Blondon AF (2017) Combining innovation in vineyard management and genetic diversity for a sustainable European viticulture. Impact 2017:28–30
Bacolla A, Tainer JA, Vasquez KM, Cooper DN (2016) Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Res 44:5673–5688
Battilana J, Lorenzi S, Moreira FM, Moreno-Sanz P, Failla O, Emanuelli F, Grando MS (2013) Linkage mapping and molecular diversity at the flower sex locus in wild and cultivated grapevine reveal a prominent SSR haplotype in hermaphrodite plants. Mol Biotechnol 54:1031–1037
Bellin D, Peressotti E, Merdinoglu D, Wiedemann-Merdinoglu S, Adam-Blondon AF, Cipriani G, Morgante M, Testolin R, Di Gaspero G (2009) Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor Appl Genet 120:163–176
Chen X, Shen Y, Zhang F, Chiang C, Pillalamarri V, Blumenthal I, Talkowski M, Wu BL, Gusella JF (2013) Molecular analysis of a deletion hotspot in the NRXN1 region reveals the involvement of short inverted repeats in deletion CNVs. Am J Hum Genet 92:375–386
Copetti D (2011) Genome analysis of transposable elements and positional cloning of disease resistance genes in grapevine. Dissertation, Università degli Studi di Udine
Di Gaspero G, Copetti D, Coleman C, Castellarin SD, Eibach R, Kozma P, Lacombe T, Gambetta GA, Zvyagin A, Cindrić P, Kovács L, Morgante M, Testolin R (2012) Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor Appl Genet 124:277–286
Dudenhöffer J, Schwander F, Töpfer R, Zyprian E (2015) Sequence analysis of loci Rpv10 and Rpv3 for resistance against grapevine downy mildew (Plasmopara viticola). Acta Hortic 1082:69–72
Eibach R, Zyprian E, Welter L, Töpfer R (2007) The use of molecular markers for pyramiding resistance genes in grapevine breeding. Vitis 46:120–124
Fechter I, Hausmann L, Daum M, Sörensen TR, Viehöver P, Weisshaar B, Töpfer R (2012) Candidate genes within a 143 kb region of the flower sex locus in Vitis. Mol Gen Genomics 287:247–259
Foria S (2015) The Rpv3 locus in grapevine: DNA variation and relevance for conventional breeding. Dissertation, Università degli Studi di Udine
Foria S, Magris G, Morgante M, Di Gaspero G (2018) The genetic background modulates the intensity of Rpv3-dependent downy mildew resistance in grapevine. Plant Breed 137:220–228
Hwang C-F, Xu K, Hu R, Zhou R, Riaz S, Walker MA (2010) Cloning and characterization of XiR1, a locus responsible for dagger nematode resistance in grape. Theor Appl Genet 121:789–799
Karaagac E, Vargas A, Andrés M, Carreño I, Ibáñez J, Carreño J, Martínez-Zapater JM, Cabezas JA (2012) Marker assisted selection for seedlessness in table grape breeding. Tree Genet Genomes 8:1003–1015
Li C, Erwin A, Pap D, Coleman C, Higgins AD, Kiss E, Kozma P, Hoffmann S, Ramming DW, Kovács LG (2012) Selection for Run1-Ren1 dihybrid grapevines using microsatellite markers. Am J Enol Vitic 64:152–155
Lobachev KS, Rattray A, Narayanan V (2007) Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front Biosci 12:4208–4220
Myles S, Chia J-M, Hurwitz B, Simon C, Zhong GY, Buckler E, Ware D (2010) Rapid genomic characterization of the genus Vitis. PLoS One 5:1–9
O'Connell LM, Ritland K (2004) Somatic mutations at microsatellite loci in Western Redcedar (Thuja plicata: Cupressaceae). J Hered 95:172–176
Pap D, Miller AJ, Londo JP, Kovács LG (2015) Population structure of Vitis rupestris, an important resource for viticulture. Am J Enol Vitic 66:403–410
Pap D, Riaz S, Dry IB, Jermakow A, Tenscher AC, Cantu D, Oláh R, Walker MA (2016) Identification of two novel powdery mildew resistance loci, Ren6 and Ren7, from the wild Chinese grape species Vitis piasezkii. BMC Plant Biol 16:170
Pathania A, Rialch N, Sharma PN (2017) Marker-assisted selection in disease resistance breeding: a boon to enhance agriculture production. In: Dubey SK, Pandey A, Sangwan RS (eds) Current developments in biotechnology and bioengineering: crop modification, nutrition, and food production. Elsevier, Amsterdam, Netherlands, pp 187–213
Regner F, Hack R, Santiago JL (2006) Highly variable Vitis microsatellite loci for the identification of Pinot Noir clones. Vitis 45:85–91
Reynolds AG (2015) Grapevine breeding programs for the wine industry. Elsevier, pp. 466
Rex F, Fechter I, Hausmann L, Töpfer R (2014) QTL mapping of black rot (Guignardia bidwellii) resistance in the grapevine rootstock ‘Börner’ (V. riparia Gm183 × V. cinerea Arnold). Theor Appl Genet 127:1667–1677
Riaz S, Tenscher AC, Rubin J, Graziani R, Pao SS, Walker MA (2008) Fine-scale genetic mapping of two Pierce’s disease resistance loci and a major segregation distortion region on chromosome 14 of grape. Theor Appl Genet 117:671–681
Schwander F, Eibach R, Fechter I, Hausmann L, Zyprian E, Töpfer R (2012) Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor Appl Genet 124:163–176
Sorensen WC, Smith EH, Smith J, Carton Y (2008) Charles V. Riley, France, and Phylloxera. Am Entomol 54:134–149
Venuti S, Copetti D, Foria S, Falginella L, Hoffmann S, Bellin D, Cindrić P, Kozma P, Scalabrin S, Morgante M, Testolin R, Di Gaspero G (2013) Historical introgression of the downy mildew resistance gene Rpv12 from the Asian species Vitis amurensis into grapevine varieties. PLoS One 8(4):e61228
Wan Y, Schwaninger HR, Baldo AM, Labate JA, Zhong GY, Simon CJ (2013) A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during neogene and quaternary climate change. BMC Evol Biol 13:141
Welter LJ, Göktürk-Baydar N, Akkurt M, Maul E, Eibach R, Töpfer R, Zyprian EM (2007) Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L.). Mol Breed 20:359–374
Zendler D, Schneider P, Töpfer R, Zyprian E (2017) Fine mapping of Ren3 reveals two loci mediating hypersensitive response against Erysiphe necator in grapevine. Euphytica 213:68
Zhang J, Hausmann L, Eibach R, Welter LJ, Töpfer R, Zyprian EM (2009) A framework map from grapevine V3125 (Vitis vinifera ‘Schiava grossa’ x ‘Riesling’) x rootstock cultivar ‘Börner’ (Vitis riparia x Vitis cinerea) to localize genetic determinants of phylloxera root resistance. Theor Appl Genet 119:1039–1051
Zyprian E, Ochssner I, Schwander F, Šimon S, Hausmann L, Bonow-Rex M, Moreno-Sanz P, Grando MS, Wiedemann-Merdinoglu S, Merdinoglu D, Eibach R, Töpfer R (2016) Quantitative trait loci affecting pathogen resistance and ripening of grapevines. Mol Gen Genomics 291:1573–1594
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgements
Grape accessions were provided by the following germplasm repositories: Consiglio per la ricerca in agricoltura e l’analisi dell’economia agraria, Italy; INRA Unité Expérimentale du Domaine de Vassal, France; Julius Kuhn Institute, Germany; Kmetijsko Gozdarski Zavod Nova Gorica, Slovenia; University of Udine, Italy; and Vivai Cooperativi Rauscedo, Italy (VCR). Chin-Feng Hwang, Dániel Pap, and Laszlo Kovács (Missouri State University) kindly provided DNA samples of V. aestivalis ‘Cynthiana’ and V. rupestris natural populations. We thank Irena Jurman, Federica Cattonaro, Nicola Ietri, and Giacomo Prete for DNA sequencing.
Funding
This research was partially funded by the EU-FP7 Innovine project (grant agreement no. 311775) and partially by the FP7-IDEAS-ERC Novabreed grant (agreement no. 294780).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
Genotypes of 307 grape accessions at InDel-25,941 and InDel-26,032, obtained by separation in capillary electrophoresis and fragment analysis with GeneMapper using default settings. (XLSX 22 kb)
ESM 2
Vitis accessions from species typical of the Midwestern US: V. rupestris, V. lincecumii, V. champinii, V. arizonica, V. aestivalis, V. longii, V. slavinii, V. doaniana, V. girdiana. (PDF 545 kb)
ESM 3
Genotypes of Vitis rupestris accessions from natural populations in Oklahoma and Missouri. (XLSX 11 kb)
ESM 4
Detailed information on InDel primers and PCR amplicons. (XLSX 12 kb)
ESM 5
Size distribution of InDels identified in the ‘PN40024’ chromosomal interval chr18:25,876,014..25,963,847 in comparison with the collinear region of BAC clone 10G13 from ‘Bianca’. Panel A shows small InDels predicted by GATK UnifiedGenotyper. Panel B shows other InDels predicted by DELLY2. (JPG 968 kb)
ESM 6
Size distribution of InDels identified in the ‘PN40024’ chromosomal intervals chr18:26,032,019..26,032,971; chr18:26,033,569..26,033,898; and chr18:26,034,098..26,034,354 in comparison with amplicons obtained from genomic DNA of ‘UD-21,076’. (JPEG 683 kb)
ESM 7
Fragment analysis chromatograms for InDel-25,941 in a set of grape accessions representative of all genotypes found in the genus Vitis. Peak height is expressed in Relative Fluorescence Units (RFU). From top to bottom: Rpv3-1homozygous, KL1270/2/14, Seyve Villard 12–358, Pinot Noir, V. rupestris du Lot. (JPG 1378 kb)
ESM 8
Fragment analysis chromatograms for InDel-26,032 in a set of grape accessions representative of the major genotypes found in the genus Vitis. Peak height is expressed in Relative Fluorescence Units (RFU). From top to bottom: Rpv3–1homozygous, Seyve Villard 12–358, Teréz, Marandi Shemakhinskii, V. acerifolia, Sauvignon Blanc, Negroamaro, Pinot Noir. (JPG 1747 kb)
ESM 9
Melting peaks of InDel-25,941 (panel A) and InDel-26,032 (panel B) amplicons. (PDF 549 kb)
ESM 10
Alignment of InDel-25,941 amplicon sequences in homozygous individuals carrying the 248-bp, 247-bp, 228-bp, and 196-bp electromorphs. (PDF 400 kb)
ESM 11
Alignment of InDel-26,032 amplicon sequences in homozygous individuals carrying the 262-bp, 244-bp, 242-bp, and 178-bp electromorphs. (PDF 398 kb)
ESM 12
Predicted ssDNA secondary structures of InDel-26,032 alleles. Highlighted in cyan are nucleotides involved in insertion/deletion mutations. (JPG 1519 kb)
ESM 13
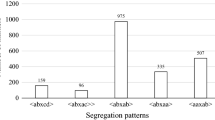
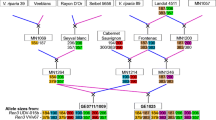
Diagram of recombinant chromosomes between InDel-25,941 or InDel-26,032 and the Rpv3–1 phenotype in eight individuals from 4221 segregant genotypes. (PDF 453 kb)
Rights and permissions
About this article
Cite this article
Foria, S., Magris, G., Copetti, D. et al. InDel markers for monitoring the introgression of downy mildew resistance from wild relatives into grape varieties. Mol Breeding 38, 124 (2018). https://doi.org/10.1007/s11032-018-0880-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0880-4