Abstract
Knowledge of the allelic variation and epistatic interaction of vernalization genes is crucial to understanding seasonal growth habit and adaptability of genetic resources in cultivated (Hordeum vulgare) and wild (Hodeum spontaneum) barley. In this research, 1398 Chinese barley accessions were characterized with molecular markers for vernalization genes HvVRN1 and HvVRN2. Different combinations of haplotypes at these two vernalization genes were present at different frequencies in wild barley, landraces and modern cultivars. All of the HvVRN1 haplotypes except HvVRN1-7 (V 1-7 ) and HvVRN1-Hs (V 1-Hs ) were identified in the Chinese barley collection chosen for this study. Haplotype V 1-8 occurred at the highest frequency in spring V 1 alleles, and V 1-1 , V 1-4 and V 1-8 had broad geographic distributions in China. Haplotypes V 1-2 and V 1-3 are restricted to modern cultivars as they were introduced from exotic germplasm. A total of 18 HvVRN1/HvVRN2 multilocus haplotypes were identified. Most of the HvVRN1/HvVRN2 haplotypes were present in materials from the Yellow-Huai River Valley and Middle-Lower Yangtze Valley ecological zones, birthplace of Chinese farming. The predominant combinations in landraces were V 1-8 /V 2 and v 1 /V 2 , but v 1 /v 2 made up the highest proportion of modern cultivars. Haplotypes V 1-5 /v 2 , V 1-10 /V 2 and V 1-9 /V 2 showed a correlation with region. Our results confirmed the presence or absence of conserved regulatory elements and the length of the first intron of HvVRN1 plays a determinant role in the vernalization requirement of barley. This study provides useful information for breeding and utilization of Chinese barley germplasm.



Similar content being viewed by others
References
Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F (2000) On the origin and domestication history of barley (Hordeum vulgare). Mol Biol Evol 17:499–510
Blake T, Blake V, Bowman J, Abdel-Haleem H (2011) Barley feed uses and quality improvement. In: Ullrich SE (ed) Barley: production, improvement, and uses. Wiley-Blackwell, Oxford, pp 522–531
Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, Martin SS, Jernstedt JA, Lagarias JC, Dubcovsky J (2014) PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc Natl Acad Sci USA 111:10037–10044
Cockram J, Chiapparino E, Taylor SA, Stamati K, Donini P, Laurie DA, O’Sullivan DM (2007) Haplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theor Appl Genet 115:993–1001
Cockram J, Hones H, O’Sullivan DM (2011) Genetic variation at flowering time loci in wild and cultivated barley. Plant Genet Res 9:264–267
Collins HM, Burton RA, Topping DL, Liao M-L, Bacic A, Fincher GB (2010) Review: variability in fine structures of noncellulosic cell wall polysaccharides from cereal grains: potential importance in human health and nutrition. Cereal Chem 87:272–282
Dai F, Nevo E, Wu D, Comadran J, Zhou M, Qiu L, Chen Z, Beiles A, Chen G, Zhang G (2012) Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA 109:16969–16973
Deng W, Casao MC, Wang P, Sato K, Hayes PM, Finnegan EJ, Trevaskis B (2015) Direct links between the vernalization response and other key traits of cereal crops. Nat Commun 6:5882
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184
Dubcovsky J, Chen C, Yan L (2005) Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breed 15:395–407
Ergon A, Hamland H, Rognli OA (2013) Differential expression of VRN1 and other MADS-box genes in Festuca pratensis selections with different vernalization requirements. Biol Plant 57:245–254
Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genom 273:54–65
Hemming MN, Peacock WJ, Dennis ES, Trevaskis B (2008) Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiol 147:355–366
Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B (2009) Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genom 282:107–117
Karsai I, Szucs P, Meszaros K, Filichkina T, Hayes PM, Skinner JS, Lang L, Bedo Z (2005) The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet 110:1458–1466
Kóti K, Karsai I, Szűcs P, Horváth C, Mészáros K, Kiss GB, Bedő Z, Hayes PM (2006) Validation of the two-gene epistatic model for vernalization response in a winter × spring barley cross. Euphytica 152:17–24
Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter × spring barley (Hordeum vulgare L.) cross. Genome 38:575–585
Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol 138:2364–2373
Lu LS (1996) Chinese barley genetics. Chinese Agricultural Press, Beijing (in chinese)
Ma DQ, Xu TW (1988) The research on classification and origin of cultivated barley in Tibet autonomous region. Sci Agric Sin 21:7–14
Oliver SN, Deng W, Casao MC, Trevaskis B (2013) Low temperatures induce rapid changes in chromatin state and transcript levels of the cereal VERNALIZATION1 gene. J Exp Bot 64:2413–2422
Preston JC, Kellogg EA (2008) Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-like genes in flowering competency and the transition to flowering. Plant Physiol 146:265–276
Saisho D, Ishii M, Hori K, Sato K (2011) Natural variation of barley vernalization requirements: implication of quantitative variation of winter growth habit as an adaptive trait in East Asia. Plant Cell Physiol 52:775–784
Sasani S, Hemming MN, Oliver SN, Greenup A, Tavakkol-Afshari R, Mahfoozi S, Poustini K, Sharifi H-R, Dennis ES, Peacock WJ (2009) The influence of vernalization and daylength on expression of flowering-time genes in the shoot apex and leaves of barley (Hordeum vulgare). J Exp Bot 60:2169–2178
Seppänen MM, Pakarinen K, Jokela V, Andersen JR, Fiil A, Santanen A, Virkajärvi P (2010) Vernalization response of Phleum pratense and its relationships to stem lignification and floral transition. Ann Bot 106:697–707
Shao QQ, Li CS, Ba SCR (1975) Origin and evolution of cultivated barley. Acta Genet Sin 2:123–128
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell Online 11:445–458
Szűcs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen TH, Hayes PM (2007) Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol Genet Genom 277:249–261
Takahashi R, Yasuda S (1971) Genetics of earliness and growth habit in barley. In: Nilan RA (ed) Barley genetics II. Washington State University Press, Pullman, pp 388–408
Tashi N, Yawei T, Xingquan Z (2013) Food preparation from hulless barley in Tibet. In: Zhang GP (ed) Advances in barley sciences. Springer, The Netherlands, pp 151–158
Trevaskis B (2010) The central role of the VERNALIZATION1 gene in the vernalization response of cereals. Funct Plant Biol 37:479–487
Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100:13099–13104
Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140:1397–1405
Trevaskis B, Hemming MN, Dennis ES, Peacock WJ (2007) The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci 12:352–357
Ullrich SE (2011) Significance, adaptation, production, and trade of barley. In: Ullrich SE (ed) Barley: production, improvement, and uses. Wiley-Blackwell, Oxford, pp 3–13
von Bothmer R, Sato K, Komatsuda T, Yasuda S, Fischbeck G (2003) The domestication of cultivated barley. In: von Bothmer R, Van Hintum Th, Knüpffer H, Sato K (eds) Diversity in Barley (Hordeum vulgare). Elsevier, Amsterdam, pp 9–27
von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59:449–467
Xu T (1987) Origin and phlylogeny of cultivated barley in China. Barley Genet 5:91–95
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004a) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109:1677–1686
Yan LL, Artem Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004b) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586
Acknowledgments
The authors are grateful to Prof. Robert McIntosh, University of Sydney, for kindly reviewing the manuscript. This research was financially supported by the National Key Technology R&D Program of the Ministry of Science and Technology (2012BAD03B01-2 and 2013BAD01B05-2), National 863 Project (2012AA101105), China Agriculture Research System (CARS-05) and the Agricultural Science and Technology Innovation Program.
Author contribution statement
DD performed the analysis of HvVRN1 and HvVRN2 haplotypes. GD, LZ, DX and ZS prepared the materials and performed DNA extraction. NT, XY and JZ phenotyped materials grown in the field and greenhouse. JZ and GG designed the experiment and prepared the manuscript. All authors have read and approved the final manuscript. Chinese barley germplasms are available for researchers and non-profit breeders by request from the National Genebank of China (http://www.cgris.net/) based on material transfer agreements.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
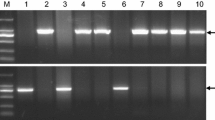
Identification of HvVRN1 and HvVRN2 alleles. Full-length Hvvrn1 (v 1 ) and V 1-6 were identified with one codominant marker. Primer pair 0.5F/R generated 1,023 bp (v 1 ) and 537 bp (V 1-6 ) products respectively. M. DL2000 DNA ladder; 1. ZDM01100 (v 1 /V 2 ); 2. ZDM09992 (v 1 /v 2 ); 3. ZDM01395 (V 1-1 /v 2 ); 4. ZDM10061 (V 1-2 /v 2 ); 5. ZDM10019 (V 1-3 /v 2 ); 6. ZDM00122 (V 1-4 /v 2 ); 7. ZDM01201 (V 1-5 /v 2 ); 8. ZDM00048 (V 1-6 /V 2 ); 9. ZDM01716 (V 1-8 /V 2 ); 10. ZDM08860 (V 1-9 /V 2 ); 11. ZDM03589 (V 1-10 /V 2 ). Supplementary material 1 (JPEG 1600 kb)
Table S1
Materials chosen for this study. +: Phenotyped by spring sown and in green house; S: spring SGH; W: winter SGH; F:facultative SGH. Supplementary material 2 (XLSX 97 kb)
Rights and permissions
About this article
Cite this article
Dondup, D., Dong, G., Xu, D. et al. Allelic variation and geographic distribution of vernalization genes HvVRN1 and HvVRN2 in Chinese barley germplasm. Mol Breeding 36, 11 (2016). https://doi.org/10.1007/s11032-016-0434-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0434-6