Abstract
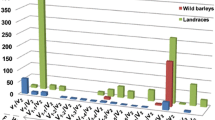
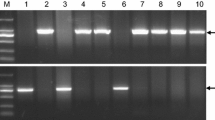
VERNALIZATION1 (VRN1) is a central regulator of the vernalization requirement, which affects the yield and geographical distribution of Triticeae crops through regulation of the flowering time. The first intron (Intron I) of VRN1 contains putative cis-elements that mediate repression of VRN1 expression. Allelic variations in Intron I are closely associated with increased VRN1 expression in vernalization-responsive accessions in the absence of vernalization. In this study, diagnostic molecular markers were developed to distinguish the main 12 known alleles, including a new allele, of barley (Hordeum vulgare) VRN1 (HvVRN1), and to investigate HvVRN1 alleles in 373 barley varieties from major agroecological regions in China. The new allele, HvVRN1-11, with a 3172-bp deletion in Intron I of HvVRN1, was identified in a Chinese barley landrace. Analysis of eight polymorphic sites flanking major variations in Intron I of HvVRN1 classified the 12 HvVRN1 alleles into five distinct groups. Compared with modern commercial barley, landraces possessed greater genetic diversity and contained the majority of HvVRN1 alleles, especially in the winter barley regions. HvVRN1 diversity in terms of allele number reduced significantly (P < 0.01) along with higher growing latitudes, but increased in accessions carrying the winter HvVRN2 allele. Wild-type HvVRN1 and the HvVRN1-8 allele showed the most widespread distribution in most regions whereas HvVRN1-1 & HvVRN1-8, and HvVRN1-5 are predominant in the Inner Mongolian Plateau spring barley region and the Northeast Plain spring barley region, respectively, with relatively higher latitudes. Compared with landraces, the frequency of HvVRN1-8 decreased while wild-type HvVRN1 increased significantly (P < 0.01) in commercial varieties. Our study classified HvVRN1 genetic diversity in a large collection of Chinese germplasm and provided a tool for identifying HvVRN1 alleles in barley breeding populations.





Similar content being viewed by others
References
Aldrich KJ, Cullis CA (1993) CTAB DNA extraction from plant tissue. Plant Mol Biol 11:128–141
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Busch MA, Bomblies K, Weigel D (1999) Activation of a floral homeotic gene in Arabidopsis. Science 285:585–587
Cockram J, Chiapparino E, Taylor SA, Stamati K, Donini P, Laurie DA, O’Sullivan DM (2007a) Haplotype analysis of vernalization loci in European barley germplasm reveals novel VRN-H1 alleles and a predominant winter VRN-H1/VRN-H2 multi-locus haplotype. Theor Appl Genet 115:993–1001
Cockram J, Mackay IJ, O’Sullivan DM (2007b) The role of double-stranded break repair in the creation of phenotypic diversity at cereal VRN1 loci. Genetics 177:2535–2539
Cockram J, Norris C, O’Sullivan DM (2009) PCR-based markers diagnostic for spring and winter seasonal growth habit in barley. Crop Sci 49:403–410
Cockram J, Hones H, O’Sullivan DM (2011) Genetic variation at flowering time loci in wild and cultivated barley. Plant Genet Res Charact Util 9:264–267
Dai F, Nevo E, Wu D, Comadran J, Zhou M, Qiu L, Chen Z, Beiles A, Chen G, Zhang G (2012) Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA 109:16969–16973
Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132:1849–1860
Devos KM, Brown JKM, Bennetzen JL (2002) Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res 12:1075–1079
Deyholos MK, Sieburth LE (2000) Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12:1799–1810
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184
Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G (1998) Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet 97:968–975
Eagles HA, Cane K, Neil V (2009) The flow of alleles of important photoperiod and vernalisation genes through Australian wheat. Crop Pasture Sci 60:646–657
Eagles HA, Cane K, Trevaskis B (2011) Veery wheats carry an allele of Vrn-A1 that has implications for freezing tolerance in winter wheats. Plant Breed 130:413–418
Fiume E, Christou P, Giani S, Breviario D (2004) Introns are key regulatory elements of rice tubulin expression. Planta 218:693–703
Fu D, Szücs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65
Gassner G (1918) Beitraege zur physiologischen Charakteristik sommer- und winter-annueller Gewaechse in besondere der Getreidepflanzen. Z Bot 10:27–476
Goncharov NP (1998) Genetic resources of wheat related species: the Vrn genes controlling growth habit (spring vs. winter). Euphytica 100:371–376
Gotoh T (1979) Genetic studies on growth habit of some important spring wheat cultivars in Japan, with special reference to the identification of the spring genes involved. Jpn J Breed 29:133–145
Greenup A, Peacock WJ, Dennis ES, Trevaskis B (2009) The molecular biology of seasonal flowering responses in Arabidopsis and the cereals. Ann Bot 103:1165–1172
Hao C, Perretant MR, Choulet F, Wang L, Paux E, Sourdille P, Zhang X, Feuillet C, Balfourier F (2010) Genetic diversity and linkage disequilibrium studies on a 3.1-Mb genomic region of chromosome 3B in European and Asian bread wheat (Triticum aestivum L.) populations. Theor Appl Genet 121(7):1209–1225
Hemming MN, Peacock WJ, Dennis ES, Trevaskis B (2008) Low temperature and daylength cues are integrated to regulated FLOWERING LOCUS T in barley. Plant Physiol 147:1–12
Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B (2009) Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genomics 282:107–117
Iwaki K, Nakagawa K, Kuno H, Kato K (2000) Ecogeographical differentiation in East Asian wheat, revealed from the geographical variation of growth habit and Vrn genotype. Euphytica 111:137–143
Iwaki K, Haruna S, Niwa T, Kato K (2001) Adaptation and ecological differentiation in wheat with special reference to geographical variation of growth habit and Vrn genotype. Plant Breed 120:107–114
Jin SB (1986) Chinese wheat cultivars and their pedigrees (1962–1982) (In Chinese). China Agriculture Press, Beijing
Jin SB (1997) Chinese wheat cultivars and their pedigrees (1983-1993) (In Chinese) China Agriculture Press, Beijing
Jung C, Műller AE (2009) Flowering time control and applications in plant breeding. Trends Plant Sci 14(10):563–573
Kamran A, Randhawa HS, Yang RC, Spaner D (2014) The effect of VRN1 genes on important agronomic traits in high-yielding Canadian soft white spring wheat. Plant Breed 133:321–326
Kapranov P, Routt SM, Bankaitis VA, de Bruijn FJ, Szczyglowski K (2001) Nodule-specific regulation of phosphatidylinositol transfer protein expression in Lotus japonicus. Plant Cell 13:1369–1382
Karsai I, Szücs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedö Z (2005) The Vrn-H2 locus is a major determinantof flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet 110:1458–1466
Kato K, Yamagata H (1988) Method for evaluation of chilling requirement and narrow-sense earliness of wheat cultivars. Jpn J Breed 38:172–186
Li C, Dubcovsky J (2008) Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J 55:543–554
Lu LS (1995) Chinese barley (In Chinese). China Agriculture Press, Beijing
Ma J, Devos KM, Bennetzen JL (2004) Analysis of LTR retrotransposon structure reveal ancient and rapid genomic DNA loss in rice. Genome Res 14:860–869
Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y (2003) WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol 44:1255–1265
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Oliver SN, Deng WW, Casao MC, Trevaskis B (2013) Low temperatures induce rapid changes in chromatin state and transcript levels of the cereal VERNALIZATION1 gene. J Exp Bot 64(8):2413–2422
Preston JC, Kellogg EA (2008) Discrete developmental roles for temperate cereal grass VERNALIZATION1/FRUITFULL-like genes in flowering competency and the transition to flowering. Plant Physiol 146:265–276
Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56:1–14
Pugsley AT (1971) A genetic analysis of the spring-winter habit of growth in wheat. Aust J Agric Res 22(1):21–31
Pumpernik D, Oblak B, Borstnik B (2008) Replication slippage versus point mutation rates in short tandem repeats of the human genome. Mol Genet Genomics 279:53–61
Sasani S, Hemming MN, Oliver S, Greenup AG, Tavakkol-Afshari R, Mahfoozi S, Poustini K, Sharifi HR, Dennis ES, Peacock WJ, Trevaskis B (2009) The influence of vernalization and daylength cues on the expression of flowering-time genes in the leaves and shoot apex of barley (Hordeum vulgare). J Exp Bot 60:2169–2178
Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, Shitsukawa N, Abe T, Kawahigashi H, Kikuchi R, Handa H, Murai K (2009) A genetic network of flowering time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J 58:668–681
Stelmakh AF (1990) Geographic-distribution of Vrn-genes in landraces and improved varieties of spring bread wheat. Euphytica 45:113–118
Stelmakh AF (1998) Genetic systems regulating flowering response in wheat. Euphytica 100:359–369
Szücs P, Skinner JS, Karsai I, Cuesta-Marcos A, Haggard KG, Corey AE, Chen THH, Hayes PM (2007) Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol Genet Genomics 277:249–261
Takahashi R, Yasuda S (1971) Genetics of earliness and growth habit in barley. In: Nilan RA (ed) Proceedings of the 2nd international barley genetics symposium. Washington State University Press, Pullman, pp 388–408
Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100:13099–13104
Trevaskis B, Hemming MN, Dennis ES, Peacock WJ (2007) The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci 12:352–357
von Zitzewitz J, Szücs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen THH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59:449–467
Wang L, He L, Li J, Zhao J, Li Z, He C (2014) Regulatory change at Physalis Organ Size 1 correlates to natural variation in tomatillo reproductive organ size. Nat Commun 5:4271. doi:10.1038/ncomms5271
Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu ZD, Dubcovsky J, Keller B (2003) Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15:1186–1197
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586
Zhang XK, Xiao YG, Zhang Y, Xia XC, Dubcovsky J, He ZH (2008) Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Sci 48:458–470
Zhuang QS (2003) Wheat improvement and pedigree analysis in Chinese wheat cultivars (In Chinese). China Agriculture Press, Beijing
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30871527, 30960196), Main Direction Program of Knowledge Innovation of Chinese Academy of Sciences (KSCX2-EW-J-22), Cooperation Project of Chinese Academy of Sciences and Tibet (XBCD-2011-019) and International Joint Project of Science & Technology Department of Sichuan Province.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Table 1
The HvVRN1 and HvVRN2 alleles among 285 Chinese barley landraces. “Code” represent codes of barley varieties in National Germplasm Bank of CAAS; “Region” indicate major barley agroecological regions in China; In column “HvVRN2”, “+” and “−” represent present HvVRN2 and absent HvVRN2, respectively (XLSX 19 kb)
Supplement Table 2
The HvVRN1 and HvVRN2 alleles among 88 Chinese barley commercial varieties. “Code” represent codes of barley varieties in National Germplasm Bank of CAAS; “Region” indicate major barley agroecological regions in China; In column “HvVRN2”, “+” and “−” represent present HvVRN2 and absent HvVRN2, respectively (XLSX 13 kb)
Supplement Table 3
PCR primers for detecting polymorphisms of the HvVRN1 and HvVRN2 locus (XLSX 9 kb)
Fig. S1
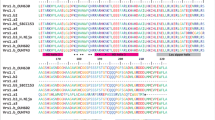
Alignment of Intron I sequence between the seven deletion alleles and the wildtype HvVRN1. Intron I deletions in HvVRN1-11 and six other alleles are flanked by short direct repeat sequences (boxed) which are indicative of non-homologous recombination following double stranded break repair (Cockram et al. 2007b). The dashed lines below denote the deleted sequence of the HvVRN1 alleles and the numbers above represent the nucleotide positions of the deletions relative to the first intron of wildtype HvVRN1 (PPTX 88 kb)
Fig. S2
Histogram of HvVRN1 frequencies in accessions carrying different HvVRN2 allele in different barley agroecological regions. S: spring barley region. W: winter barley region. –HvVRN2: HvVRN2 absent. +HvVRN2: HvVRN2 present (DOCX 13 kb)
Supplement Table 4
Typical sowing and heading dates for the different Chinese agroecological regions (XLSX 8 kb)
Rights and permissions
About this article
Cite this article
Zhang, C.H., Xu, D.A., Zhao, C.H. et al. Identification and distribution of VERNALIZATION1 alleles in Chinese barley (Hordeum vulgare) germplasm. Mol Breeding 35, 162 (2015). https://doi.org/10.1007/s11032-015-0346-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0346-x