Abstract
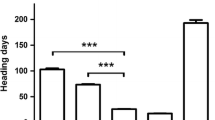
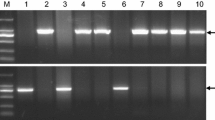
The epistatic interaction of alleles at the VRN-H1 and VRN-H2 loci determines vernalization sensitivity in barley. To validate the current molecular model for the two-locus epistasis, we crossed homozygous vernalization-insensitive plants harboring a predicted “winter type” allele at either VRN-H1 (Dicktoo) or VRN-H2 (Oregon Wolfe Barley Dominant), or at both VRN-H (Calicuchima-sib) loci and measured the flowering time of unvernalized F2 progeny under long-day photoperiod. We assessed whether the spring growth habit of Calicuchima-sib is an exception to the two-locus epistatic model or contains novel “spring” alleles at VRN-H1 (HvBM5A) and/or VRN-H2 (ZCCT-H) by determining allele sequence variants at these loci and their effects relative to growth habit. We found that (a) progeny with predicted “winter type” alleles at both VRN-H1 and VRN-H2 alleles exhibited an extremely delayed flowering (i.e. vernalization-sensitive) phenotype in two out of the three F2 populations, (b) sequence flanking the vernalization critical region of HvBM5A intron 1 likely influences degree of vernalization sensitivity, (c) a winter habit is retained when ZCCT-Ha has been deleted, and (d) the ZCCT-H genes have higher levels of allelic polymorphism than other winterhardiness regulatory genes. Our results validate the model explaining the epistatic interaction of VRN-H2 and VRN-H1 under long-day conditions, demonstrate recovery of vernalization-sensitive progeny from crosses of vernalization-insensitive genotypes, show that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity, and provide molecular markers that are accurate predictors of “winter vs spring type” alleles at the VRN-H loci.





Similar content being viewed by others
References
Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427:164–167
Ben-Naim O, Eshed R, Parnis A, Teper-Bamnolker P, Shalit A, Coupland G, Samach A, Lifschitz E (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46:462–476
Danyluk J, Kane ND, Breton G, Limin AE, Fowler DB, Sarhan F (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132:1849–1860
Dubcovsky J, Chen C, Yan L (2005) Molecular characterization of the allelic variation at the VRN-H2 vernalization locus in barley. Mol Breed 15:395–407
Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G (1998) Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor Appl Genet 97:968–975
Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L (2006) Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol Biol 60:469–480
Fu D, Szűcs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131:1855–1867
Hayes PM, Blake T, Chen THH, Tragoonrung S, Chen F, Pan A, Liu B (1993) Quantitative trait loci on barley (Hordeum vulgare) chromosome 7 associated with components of winterhardiness. Genome 36:66–71
Hayes PM, Corey AE, Dovel R, Karow R, Mundt C, Rhinart K, Vivar H (2000) Registration of Orca barley. Crop Sci 40:849
Iwaki K, Haruna S, Niwa T, Kato K (2001) Adaptation and ecological differentiation in wheat with special reference to geographical variation of growth habit and Vrn genotype. Plant Breed 120:107–114
Kane NA, Danyluk J, Tardif G, Ouellet F, Laliberté J, Limin AE, Fowler DB, Sarhan F (2005) TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant Physiol 138:2354–2363
Karl BN, Hugh BN (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. (Distributed by the author)
Karsai I, Hayes PM, Kling J, Matus IA, Mészáros K, Láng L, Bedő Z, Sato K (2004) Genetic variation in component traits of flowering time in Hordeum vulgare subsp. spontaneum accessions characterized in controlled environments. Crop Sci 44:1622–1632
Karsai I, Mészáros K, Láng L, Hayes PM, Bedő Z (2001) Multivariate analysis of traits determining adaptation in cultivated barley. Plant Breed 120:217–222
Karsai I, Mészáros K, Szűcs P, Hayes PM, Láng L, Bedő Z (2006) The influence of photoperiod on the Vrn-H2 locus (4H) which is a major determinant of plant development and reproductive fitness traits in a facultative × winter barley (Hordeum vulgare L.) mapping population. Plant Breed 125:468–472
Karsai I, Szűcs P, Mészáros K, Filichkina T, Hayes PM, Skinner JS, Láng L, Bedő Z (2005) The Vrn-H2 locus is a major determinant of flowering time in a facultative × winter growth habit barley (Hordeum vulgare L.) mapping population. Theor Appl Genet 110:1458–1466
Kooiker M, Airoldi CA, Losa A, Manzotti PS, Finzi L, Kater MM, Colombo L (2005) BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17:722–729
Kóti K, Karsai I, Szűcs P, Horváth C, Mészáros K, Kiss GB, Bedő Z, Hayes PM (2006) Validation of the two-gene epistatic model for vernalization response in a winter × spring barley cross. Euphytica 152:17–24
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lasa JM, Igartua E, Ciudad FJ, Codesal P, Garcia EV, Gracia MP, Medina B, Romagosa I, Molina-Cano JL, Montoya JL (2001) Morphological and agronomical diversity patterns in the Spanish barley core collection. Hereditas 135:217–225
Laurie DA, Pratchett N, Bezant JH, Snape JW (1995) RFLP mapping of five major genes and eight quantitative trait loci controlling flowering time in a winter 3 spring barley (Hordeum vulgare L.) cross. Genome 38:575–585
Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiol 138:2364–2373
Michaels SD, Amasino RM (1999) Flowering locus C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y (2003) WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol 44:1255–1265
Reinheimer JL, Barr AR, Eglinton JK (2004) QTL mapping of chromosomal regions conferring reproductive frost tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 109:1267–1274
Robson F, Costa MMR, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G (2001) Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J 28:619–631
Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W (2000) Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol 42:899–913
Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14:2527–2537
Skinner JS, Szűcs P, von Zitzewitz J, Marquez-Cedillo L, Filichkin T, Stockinger EJ, Thomashow MF, Chen THH, Hayes PM (2006) Mapping of barley homologs to genes that regulate low temperature tolerance in Arabidopsis. Theor Appl Genet 112:832–842
Szűcs P, Karsai I, von Zitzewitz J, Mészáros K, Cooper LLD, Gu YQ, Chen THH, Hayes PM, Skinner JS (2006) Positional relationships between photoperiod response QTL and photoreceptor and vernalization genes in barley. Theor Appl Genet 112:1277–1285
Takahashi R, Yasuda S (1971) Genetics of earliness and growth habit in barley. In: Nilan RA (ed) Barley genetics II. Proceedings of the second international barley genetics symposium. Washington State University Press, Pullman, pp 388–408
Takatsuji H (1998) Zinc-finger transcription factors in plants. Cell Mol Life Sci 54:582–596
Tottman DR, Makepeace RJ (1979) An explanation of the decimal code for the growth stages of cereals, with illustrations. Ann Appl Biol 93:221–234
Tranquilli GE, Dubcovsky J (2000) Epistatic interactions between vernalization genes Vrn-A m 1 and Vrn-A m 2 in diploid wheat. J Hered 91:304–306
Trevaskis B, Bagnall DJ, Ellis MH, Peacocck WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100:13099–13104
Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140:1397–1405
von Zitzewitz J, Szűcs P, Dubcovsky J, Yan L, Pecchioni N, Francia E, Casas A, Chen THH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59:449–467
Wolfe RI, Franckowiak JD (1991) Multiple dominant and recessive genetic marker stocks in spring barley. Barley Genet Newsl 20:117–121
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004a) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109:1677–1686
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004b) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100:6263–6268
Acknowledgments
We wish to thank Douglas L. Heckart, John L. Helgesson, and Lauren A. Osborn for their technical assistance. This work was supported by the National Science Foundation Plant Genome Research Program (DBI 0110124) and the United States Barley Genome Project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
Szűcs, P., Skinner, J.S., Karsai, I. et al. Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol Genet Genomics 277, 249–261 (2007). https://doi.org/10.1007/s00438-006-0195-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0195-8