Abstract
Over the past year, owing to the emergent demand for the search for potential COVID-19 therapeutics, identifying alternative candidates from biological sources is one of the sustainable ways to reinforce the drug discovery process. Marine macroalgae have numerous advantages because of the richest availability of underexploited bioactive compounds. Polyphenolic compounds like phlorotannins obtained from brown macroalgae are reported as proven antiviral and immunostimulatory agents. Thus, the present study evaluated the possibility of phlorotannins as antagonists to the multiple target proteins essential for SARS-CoV-2 replication. Twenty different types of potent phlorotannins were targeted against druggable target proteins, viz., 3CLpro, RdRp, and Spro using AutoDock molecular docking, drug-likeness were assessed by ADMET profiling (QikProp module). Further, validated with 200 ns molecular dynamics (MD) simulation (Desmond module) for the top-ranked phlorotannins based on docking binding affinities. Among the twenty phlorotannins studied, eckol hexacetate, phlorofucofuroeckol, fucofuroeckol, and bifuhalol-hexacetate showed significant binding affinities across the selected targets. Besides, MD simulations highlighted Glu166, Gln189, Cys145, and Thr190 tetrad as potential interaction sites to inhibit 3CLpro's activity. Moreover, phlorotannins were confirmed to be druglike, with no major deviation observed in ADMET-profiling. Hence, phlorotannins could be therapeutic candidates against SARS-CoV-2. However, further investigations are needed to prove its efficacy as an antiviral agent. Conclusively, this study may envisage that the novel finding could notably impact the advancement of antiviral interventions for COVID-19 in the near future.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
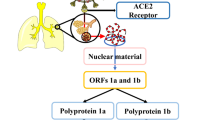
Coronavirus are zoonotic viruses, which have gained much attention nowadays because of its ability to cause infection across species. Coronavirus is a positive-sense single-stranded RNA virus belonging to the coronaviridae family and consists of four genera: alpha, beta, gamma and delta. Among which Alphacoronavirus and Betacoronavirus infect animals, whereas Gammacoronavirus infects avian species and Deltacoronavirus infects both mammals and avian species [1]. Among these, Betacoronaviruses are more commonly explored and the novel coronavirus (2019-nCoV) comes under this category which originated from Wuhan, China, in December 2019, which has created a pandemic situation and spread across more than 200 countries. The etiology of COVID-19 disease is a SARS-CoV-2 (novel severe acute respiratory syndrome-related coronavirus) virus (also known as 2019-nCoV), whereas phylogenetic analysis has a close resemblance with past epidemic human SARS virus originated from bats [2]. The SARS-CoV-2 consists of the largest genome among all RNA viruses ranging between 27 and 32 kb with four main structural proteins, namely spike, membrane, envelope and nucleocapsid proteins that helps in the recognition and attachment of the virus to the host receptor [3]. Among these proteins, three main proteins play a prominent role in the life cycle of this virus, which includes the spike glycoprotein (Spro), 3CL protease (3CLpro) and RNA-dependent RNA polymerase (RdRp). The spike glycoprotein plays a crucial role by helping the virion recognize and bind to human Angiotensin-Converting Enzyme 2 (hACE2) protein which is the most important receptor for binding SARS-CoV-2. That is mediated by two important functional units, namely the N and C terminal subunits. N-terminal subunit forms the globular head of spike protein and the C-terminal S2 region forms the stalk of the protein. These two subunits help the virus recognize and fuse with the host cell receptor hACE2 [4]. The SARS-CoV-2 consists of two overlapping open reading frames (ORF1a and ORF1b) which codes for two different polyproteins pp1a and pp1ab processed by 3CL protease and papain-like protease resulting in 16 different mature nonstructural proteins. Thus, 3CL protease plays a chief role in the replication of the SARS-CoV-2 within the host [5]. Inside the host, the elongation of the viral genome is carried out by the formation of replication and transcription complex (RTC). RTC is formed by uniting several NSPs (non-structural proteins) that are released from pp1ab cleavage. In this multifunctional elongation mechanism, the major role is contributed by nsp12, also called as RdRp [6]. The salient role played by these three proteins in attachment and replication has gained much attention in using them as a target for drug delivery. In recent days therapies for COVID-19 mainly focuses on either drug targeting the virus or the action of drugs acting on the human immune system to improve human health. However, foresaid strategies are falling short because of the emergence of new mutated viral strains with altered antigenicity [7].
The present crisis clearly indicates the urgent need for developing novel antiviral therapeutics that are effective against mutated SARS-CoV-2 viral strains. Perhaps, the development of the antiviral candidates, molecules with novel functional groups play a critical role in the structure and effectiveness of drugs [8, 9].
However, in-silico approaches show a significant role in the development of efficient antiviral candidate molecules. Furthermore, the use of in-silico techniques is neither laborious nor expensive and can be exploited for widely anticipated interactions between bioactive chemicals with the biological life systems.
Over the past years, identifying alternative and practical therapeutic candidates from biological sources has been a sustainable way to reinforce the drug discovery process against various new emerging diseases. Numerous potent bioactive candidates have been discovered from mining marine natural products with pharmacological potential, the marine pharmacopeia remains an invaluable resource of biological and chemical diversity [41,42,43]. Seaweeds are underexploited and an excellent source of bioactive compounds. Seaweeds contain diverse metabolites as natural defense compounds that equip them to survive in extreme environmental conditions. Different algae-derived bioactive compounds such as sulfated polysaccharides, polyphenols (phlorotannins), carotenoids, essential fatty acids, vitamins, amino acids, minerals, and dietary fibers are being used against many non-communicable and communicable diseases, including microbial and viral infections [10, 11]. Marine polyphenols are a group of organic compounds that are found exclusively and abundantly in brown macroalgae. Several reports have proved that the consumption of polyphenols would aid the regulation of chronic diseases, metabolism, body weight, etc. Besides, marine polyphenols are extensively reported for pharmacological properties like antioxidant, anticancer, antiviral, antibacterial, and anti-inflammatory properties [12]. Therefore, applications of marine-derived bioactives display a hopeful way to develop novel therapeutics to combat various diseases.
Phlorotannins are one of the major secondary metabolites which are exclusively found in marine brown algae. Phlorotannins are a diverse group of polyphenols in terms of structure and degree of polymerization, and as a result, they possess a broad spectrum of potential bioactivities [13, 14]. Interestingly, phlorotannins have also been reported as antiviral agents against SARS-CoV (a coronavirus) [15], influenza A viral strains (H1N1 and H9N2) [16], human papillomavirus (HPV—HPV16PVs and HPV18PVs) [17], human immunodeficiency virus type-1 (HIV-1) [18], porcine epidemic diarrhea virus (PEDV) (a coronavirus) [19], viral hemorrhagic septicemia virus (VHSV) [20], and murine norovirus (MNV) [21, 22]. These salient features of phlorotannins have added more attention to upcoming researchers and scientists to explore the insights of these medicinal-valued compounds for treating the COVID-19 since there are no suitable drugs available. Therefore, it is important to evaluate the inhibitory effects of each phlorotannin isolated from brown macroalgae on major target proteins such as 3CLpro, RdRp and Spro of SARS-CoV-2.
Therefore, the present study is focused on evaluating effective antiviral candidate molecules from selected phlorotannins (20 Nos) with three main targets of SARS-CoV-2 by molecular docking studies to explore the best binding pose with minimum binding energy and further validated using molecular dynamics simulation to evaluate the structural stability, interactions and constancy of macromolecule interactions with the ligand. Additionally, the best suitable drug candidate was determined using in-silico ADMET profiling, which extends the details on the absorption, distribution, metabolism, excretion, and toxicity of drug molecules that play an essential role in the initial screening of drugs for clinical phases. Conclusively, the results were compared with the standard COVID-19 antiviral drugs to validate the anticipated phlorotannins.
Methods and computational details
Protein targets
Three major functional proteins, 3-Chymotrypsin-like protease (3CLpro) (PDB ID: 6Y2E), RNA-Dependent RNA Polymerase (RdRp) (PDB ID: 6M71) and spike glycoprotein (Spro) (PDB ID: 6VYB) that plays a crucial role in infection and proliferation of SARS-CoV-2 were selected as druggable targets. The protein structures of the proteins were retrieved from the Protein Data Bank (PDB). Protein structures were pre-processed for further docking studies to warrant the accuracy of the structure by removing non-protein atoms and segregation of multimeric chain protein to single-chain units by pyMOL and Discovery Studio tools (Dassault System BIOVIA) as described in the previous report [23].
Ligand preparation
The 3D Structure Data File (SDF) of twenty marine brown macroalgal phlorotannin ligand molecules were obtained from the PubChem database as illustrated in Supplementary Figure S1. For ligands with only 2D SDF formats, the 3D structures were derived from the Discovery Studio 2020 and PyMol visualization tool (Schrödinger, Inc.). Concurrently, the protein targets were also docked with the repurposed antiviral drugs used against SARS-CoV-2, such as dexamethasone, remdesivir, favipiravir and MIV-150 (anti-retroviral) to compare their efficacy and to evaluate the hostile effect of phlorotannin ligands.
Molecular docking
AutoDockTools 1.5.6 (ADT) (Scripps Research, US) with the extension suite to the Python Molecular Viewer of MGL tools with Cygwin program was employed for molecular docking of ligand molecules with protein targets of SARS-CoV-2. Prior to docking, the ligand and protein were pre-processed with water deletion, polar addition, and merging of non-polar hydrogen bonds. Concurrently, the Kollman charges were fitted computationally to derive the electrostatic potential of amino acid of the protein prior to docking by ADT. Later, PDBQT format files were generated to input grid parameter file (GPF) and Docking Parameter File (DPF). The designated GPF was outlined with Lamarckian Genetic Algorithm (LGA) with 2.5 × 104 energy assessment. Eventually, the DPF was assigned with a total of 50 individualistic runs to enhance and determine the unique binding pocket of the target protein with higher interaction efficiency. Finally, the docking action was initiated using the Cygwin platform. The ideal docking complex was investigated by the confirmation with the least binding energy and lowest RMSD value obtained from ADT. The docking score and Ki (inhibition concentration) data were retrieved in a log file (DLG) from Cygwin. Further, the best interaction complex was classified based on the lowest binding energy, hydrogen bonding, hydrophobic interactions, and other favorable interactions of complex molecules analyzed in the Discovery Studio platform.
Molecular dynamics simulation
Molecular Dynamics (MD) simulations were performed using the Desmond tool in the Maestro platform of Schrödinger software for the top-ranked ligands, namely eckol hexacetate, phlorofucofuroeckol B, fucofuroeckol, bifuhalol hexacetate and phlorofucofuroeckol, which had shown the least binding scores from molecular docking analysis. The protein was preprocessed, optimized with H-bond, and minimized with OPLS3e force field. Using system builder, the protein and ligand complex was solvated in the 3D orthorhombic box with a buffer volume of 853,903 Å3 and distance 10 Å, respectively, to build a hydration model (TIP3P water model). The MD simulation time was carried out up to 200 ns under NPT ensemble (constant number of atoms (N), constant pressure (P) & temperature (T)) where the model system was relaxed prior to simulation using the default Desmond settings. The outputs of the simulation results were analyzed using a simulation interactions diagram tool that provided data on complex macromolecule-ligand properties of RMSF (root mean square fluctuation) to calculate the flexibility of individual residue. The RMSD (root mean square distance) compares the prediction error in disparate models of specific data sets and protein–ligand contacts with the timeline.
Molecular mechanics-generalized born surface area (MM-GBSA)
The docking posture of the ligand was rescored using Prime's MM-GBSA approach. These poses were used as inputs for minimizing the energy of protein–ligand complexes (PL), free proteins (P), and free ligands (L). The binding free energy dGbind was calculated using the equation as follows:
The MM-GBSA uses Prime VSGB solvation model with applied force field of OPLS4 for energy minimization (Schrödinger Release 2021–3: Prime, Schrödinger, LLC, New York, NY, 2021).
Prediction of pharmacokinetics
Pharmacokinetics analyses of the marine macroalgal phlorotannin ligand molecules were prognosticated by applying compartmental physiological features based on a mathematical equation using the statistical approach by QikProp module in the platform of Maestro of Schrodinger’s software. The major intrinsic parameters for the prediction of maximum drug likelihood elucidations were validated based on Lipinski's rules, crossing the blood–brain barrier, and molecular weight of small molecules.
Results and discussion
The lack of therapeutics against COVID-19 highlights the urgent need to virtually screen alternative bioactives with antiviral properties to inhibit the SARS-CoV-2 entry and replication. Phlorotannins, a class of polyphenols mainly found in marine brown algae, are investigated for potential inhibition of three major druggable protein targets of SARS-CoV-2 such as 3CLpro, RdRp, and Spro, which play a crucial role in the viral replication process. Indeed, diverse phlorotannins have been widely reported as antiviral, antioxidant, antimicrobial, antidiabetic, anticancer, radioprotective, neuroprotective agents [24, 25]. Thus, the present study evaluates twenty different types of phlorotannins as probable antagonists against the SARS-CoV-2 target proteins using molecular docking. The ADMET profiling was done for the twenty phlorotannins to check for drug suitability. The top phlorotannins with the best binding scores were further validated to determine the stability of the ligand–protein complex with molecular dynamics simulation analysis.
Molecular docking: screening of potent inhibitors for SARS-CoV-2 major target proteins
The preparatory assessment of the antagonistic effect of twenty-brown algal bioactive polyphenols against major proteins of SARS-CoV-2 was evaluated based on binding energy efficiency. For a virtual docking process, an implied ligand should fit the active site of the protein target. Thus, the main aim relies upon the identification of the most stable binding interaction mode. The affinity of the selected ligands to form durable interactions inside targets' pockets is represented by their binding energy values. Strong binding of the ligand–protein complex is indicated by a low binding energy value and vice versa.
The comparative investigation of the binding energy of the ligand molecules with three target proteins is summarized in Table 1. To our surprise, the hampering action on 3CL protease was highly coherent with greater binding energy by all 20 algal bioactive phlorotannins. Furthermore, all the ligands had the closest interaction with 3CLpro than other targets such as RdRp and Spro. The maximum score of lowest binding energy in 3CLpro was observed in eckol hexacetate with −9.10 kcal/mol. Similarly, the lowest binding energy for RdRp and Spro was noticed in dioxinodehydroeckol (−5.57 kcal/mol) and bifuhalol hexacetate (−6.24 kcal/mol), respectively. Interestingly, the binding efficiency of eckol hexacetate with RdRp and Spro remains at the top three leading positions confessing the efficacy of the antagonistic nature of ligand molecule. Similarly, phlorofucofuroeckol A was noticed to show low binding energy in all the three protein targets with top five positions. Moreover, other top-six phlorotannins like difucol hexaacetate, fucofuroeckol, phlorofucofuroeckol B, diphlorethol pentaacetate, and bifuhalol hexaacetate showed better binding energy scores respective to all the three targets. In our previous report, the phlorotannins eckol and trifucol have also demonstrated good binding energy scores with 3CLpro (−8.19 and −6.36 kcal/mol) and Spro (−4.10 and −7.57 kcal/mol), respectively. In comparison with standard experimental antiviral drugs used for COVID-19, dexamethasone was showed the lowest binding energy with 3CLpro (−7.19 kcal/mol) followed by remdesivir (−6.82 kcal/mol) and favipirivir (−4.01 kcal/mol). Similarly, dexamethasone was showed the lowest binding energy with Spro (−4.35 kcal/mol) followed by remdesivir (−3.87 kcal/mol) and favipirivir (−3.53 kcal/mol). Further, the results were also compared with anti-retroviral drug MIV-150 (prepared from marine algal source), which resulted in low binding energy with 3CLpro (−7.57 kcal/mol) and Spro (−5.07 kcal/mol). The phlorotannin ligands' good binding activity with SARS-CoV-2 target proteins might be due to the presence of hydroxyl moieties in their frame structures as shown in Fig. 2 [26].
The current work is in concordance with the recent study on phlorotannins as potential antiviral candidates against two SARS-CoV-2 protein targets. In which, out of eleven phlorotannin ligands, lower binding energy toward RBD/ACE2 were observed with dieckol (−7.40 kcal/mol), 8,8-Bieckol (−7.25 kcal/mol), phlorofucofuroeckol B (−6.93 kcal/mol), phlorofucofuroeckol A (−6.67 kcal/mol), and 6,6-Bieckol (−6.65 kcal/mol), and were comparatively better than the standard antiviral drugs like remdesivir (−6.53 kcal/mol), chloroquine (−5.23 kcal/mol), and hydroxychloroquine sulfate (−5.18 kcal/mol). Similarly, lower binding energy toward Spike/TMPRSS2 were observed with 6,6-Bieckol (−9.17 kcal/mol), dieckol (−9.03 kcal/mol), phlorofucofuroeckol B (−8.51 kcal/mol), diphloroethohydroxycarmalol (−8.31 kcal/mol), 7-Phloroeckol (−8.28 kcal/mol), which were better than the standard drugs remdesivir (−8.01 kcal/mol), chloroquine (−5.73 kcal/mol), and hydroxychloroquine sulfate (−5.92 kcal/mol) [27]. Similar results were also reported by Gentile et al. where phlorotannins ranked top among 14,064 marine natural products when screened against SARS-CoV-2 main protease (Mpro) using pharmacophore model, and molecular docking approaches. In which phlorotannins Heptafuhalol A, Phlorethopentafuhalol B, Pseudopentafuhalol C, Phlorethopentafuhalol A, Hydroxypentafuhalol A, Pentaphlorethol B, 8,8’-Bieckol, 6,6’-Bieckol, Dieckol, and Pentaphlorethol A exhibited least free binding energies [44]. The docked macromolecule (protein) and small molecule (ligand) are called complex molecules, and their interaction is shown in three-dimensional and two-dimensional pictorial graphs in Figs. 1, 2. The 3D and 2D interaction graphs represent electrophilic interactions introduced from hydroxyl group interaction by the target proteins with hydrogen bonds. The hydrogen bond represents the most-closest interaction of the complex molecules with bond length extends 2.5 to 3.5 Å. In addition, the hydrophobic and Vanderwaal interactions support the contact with the distance of 3.5 Å to 4.5 Å. The overall possible amino acid interaction of all 20 ligands with 3CLpro are Glu166, Ser144, Leu141, His164, Asn142, Met49, Met165, Cys44, His41, Pro168, Thr26, Cys145, Gly143, Thr25, Ser46, Phe140, Met49, Leu167, Leu27, Thr24 in the range of 0.3 to 5.82 Å distance; for spike glycoprotein as N501, R403, E406, K417, Q409, R408, G496, S494, Q493, Y453, R454, Y505, F497, D405 in the range of 1.73 to 5.98 Å distance; and for RdRp as L387, V398, A382, A383, S384, L271, T324, V330, L329, L270, Ser709, Lys714, Lys780, Asn781, Ala706, Gly774, Tyr129, Thr710, Pro328, Ala379, Met666, Thr324, Phe396, Val675, Ser664, Arg349, R555, K545, A547, S549, F441, H439, R836, I548, R858, D845, Pro322, Pro323, Thr319, Ile266, Ser255, Phe321, Lys267, Trp268, Tyr265, Val320, Asp135, Tyr32, Lys47, Ser784, Gly774, Ser325, Gly327, Leu329, Pro677, Leu460, Asn459, Arg457, Lys276, Thr394, Cys395, Ser197, Leu388, Leu389, Pro421, Ala771, Gln773, Lys714, Lys780, Asp711 in the range of 1.75 to 5.69 Å distance. The Glu166 & Gln189 dyad was conserved in all polyphenolic compounds against 3CLpro, whereas N501, R403, E406, G496, and Y453 penta-complex amino acid was showed conserved across all phlorotannin compounds against Spro, and no significant conserved regions were found in RdRp (Table 2). Similar amino acid interaction patterns were reported for SARS-CoV-2 3CLpro with the novel quinoline identified from medicinal plant Diplocyclos palmatus [28].
Three-dimensional representation of molecular interaction of marine brown algal phlorotannins with SARS-CoV-2 target proteins. 3CLpro with a Eckol hexaacetate & b Diphlorethol pentaacetate; Spro with c Bifuhalol hexaacetate & d Eckol hexaacetate; RdRp with e Dioxinodehydroeckol & f Phlorofucofuroeckol B
Two-dimensional structure depiction of molecular interaction of marine brown algal phlorotannins with SARS-CoV-2 target proteins. 3CLpro with a Eckol hexaacetate & b Diphlorethol pentaacetate; Spro with c Bifuhalol hexaacetate & d Eckol hexaacetate; RdRp with e Dioxinodehydroeckol & f Phlorofucofuroeckol B
The relative free binding energies of each complex molecule have been calculated as prime energy calculation using the Prime MM-GBSA module in Schrödinger software. MM-GBSA calculations were done in order to improve the ranking of the ligands and to determine forecast binding energies. The calculated values were approximate binding free energies, with a higher negative value indicating a stronger binding.
In the present study, the top six complex molecules have shown promising interactions with 3CLpro shown in the range of −15.41 to −52.14 kcal/mol free energies, RDRP complex with −26.15 to −39.07 kcal/mol and Spro between −37 and −46.79 kcal/mol as listed in Table 3. However, the wide range of free energies resulted in the present study, the highest free energy was observed in Phlorofucofuroeckol B (−15.41 kcal/mol) and the least binding energy was observed in Phlorofucofuroeckol A (−52.14 kcal/mol) among the three main targets.
An increasing number of reports on phlorotannins as broad-spectrum antiviral agents against SARS-CoV-1 [9], PEDV [22], influenza virus [10], HIV-1 [12], HPV [11], VHSV ([14]), and MNV [12, 16] implies their importance as alternative natural therapeutic candidates. Furthermore, since both SARS-CoV-2 and SARS-CoV-1 have very similar 3CLpro, therefore phlorotannins which were reported as 3CLpro inhibitors earlier, may have the possibility to interact with 3CLpro of SARS-CoV-2 also [29].
Thus, the current study was designed based on the previous work [15], in which it was reported that nine phlorotannins (phloroglucinol, eckol, dieckol, triphloretol A, 2-phloroeckol, dioxinodehydroeckol, phlorofucofuroeckol A, fucodiphloroethol G, and 7-phloroeckol) extracted from Ecklonia cava (edible brown algae) that showed potent inhibitory activity against SARS-CoV-1 3CLpro in a dose-dependent with competitively no toxicity. In addition, among these phlorotannins, dieckol was reported to have the most potent 3CLpro trans/cis-cleavage inhibitory effects. Moreover, molecular docking experiments also strengthened the inhibitory effect of dieckol against SARS-CoV 3CLpro with a lowest binding energy of −11.51 kcal/mol compared to other phlorotannins [15].
Interestingly, dieckol and diphlorethohydroxycarmalol, have also been reported for HIV-1 reverse transcriptase (RT) and HIV-1 protease inhibition. Also, dieckol and 6,6-Bieckol hindered the HIV penetration, syncytia formation, viral replication, and lytic effects in cells in a dose-dependent manner [18, 30]. Similarly, phlorotannins phlorofucofuroeckol, dieckol, 7-Phloroeckol, and eckol have also been reported for their inhibitory activity against coronavirus—Porcine Epidemic Diarrhea Virus (PEDV) by strong interaction with spike protein resulting in preventing the viral attachment to the cells, inhibition of hemagglutination [19]. Furthermore, in a recent report by [25], the marine seaweed compounds library was screened by HTVS (High-Throughput Virtual Screening) against the SARS-CoV-2 spike protein RBD. Out of 1110 compounds screened from the library, the compound dieckol (phlorotannin) was reported to be the potent spike protein RBD binder based on screening by SP docking, XP docking [31]. Similarly, Adegboyega et al., (2021) had reported inhibitory activity of five antiviral drugs (remdesivir, lopinavir, dieckol, ritonavir, and chloroquine) against SARS-CoV-2 target proteins (RdRp, 3CLpro, spike protein, PLpro, and Mpro) by molecular docking analysis in which phlorotannin dieckol comparatively docked at the binding site of all target proteins better than other antivirals with binding scores of −9.3, −9.3, −8.5, −9.6, and −9.2 kcal/mol, respectively [32].
The current study exhibited that eckol hexaacetate could dock with the SARS-CoV-2 target protein binding site with additional hydrogen, thus resulting in the higher binding energy compared to other screened ligands. Therefore, eckol hexaacetate may reduce the activity of SARS-CoV-2 by blocking the active site of the target proteins in the groove. Thus, the results imply that phlorotannins like eckol hexacetate, phlorofucofuroeckol A, bifuhalol hexacetate, dioxinodehydroeckol, difucol hexaacetate, fucofuroeckol, phlorofucofuroeckol B, eckol, trifucol, and diphlorethol pentaacetate could be a better natural alternative therapeutic option considering the least binding energy scores in comparison to standard antiviral drugs. However, to the best of our knowledge, this is the first report on molecular docking and dynamics simulation analyses for phlorotannins binding affinity on multitargets RdRp, Spro, and 3CLpro of SARS-CoV-2.
Hostile effects at functional domains
The irreversible interaction of ligand molecules on functional domains of target protein causes severe loss of activity. Herein, the binding interaction of the various brown algal phlorotannin on the functional domain of the RdRp, Spro and 3CL protease were investigated to determine the hostile effects. The nsp12 active domain is the central core component of the catalytic subunit of RdRp, which is entailed for the RNA polymerizing activity [33]. The stretches of amino acid residues from 398 to 932 in RdRp configures the hydrophobic pockets that are involved in the active functionalization of RNA polymerization [33]. Kirchdoerfer et al., (2019) categorized the nsp12 region of the polymerase domain as finger domain (amino acids 398 to 581; 628–687), palm domain (amino acids 582–627; 688–815) and, thumb domain (amino acids 816–919). The initial stretches of the amino acids (1–397) are nidoviral (coronavirus family) amino-terminal sequences [35]. The screening of binding interaction of the brown algal phlorotannin in the foresaid three functional domains of RdRp were analyzed to determine the effective antagonistic effect. Surprisingly, the interaction of the different brown algal phlorotannins were highly conserved at a specific region of the distinct domains of the polymerase. Especially nine out of 20 compounds showed the amino acid interaction only with the Nidoviral (1 to 397) N-terminal extension. Likewise, the triphloroethol A and 7-Phloroeckol had shown the amino acid interaction only at the palm domain. The hostile effect of the brown algal phlorotannin at different functional domains of the RdRp is given in Table 4.
Similarly, the antagonistic effects of brown algal phlorotannin were also investigated with the receptor-binding domain of the spike protein. Mainly, the interactions that occurred at the specific Receptor Binding Motif (RBM) sites were highly focused, which was involved in the attachment of angiotensin-converting enzyme 2 (ACE2) of the human lung cells. The core region of the RBM was localized at the particular stretches of the amino acid sequences in between 438 and 506 in the spike protein of SARS-CoV-2 [36]. Interestingly, all of the brown algal phlorotannins was prevalently exhibited binding interactions at the exact site of the RBM. The amino acid interaction occurred between 403 and 505 virtually in every individual compound, which is identical to the RBM of the spike protein. The results clearly showed that the brown algal phlorotannins were eventually targeting the active sites of the receptor-binding domain of the SARS-CoV-2.
Similar analyses were performed on the 3CL protease to determine the hindering interaction at the catalytic functional domains. The 3CL protease consisted of three different functional domains as antiparallel (amino acids 8–101), β-barrel (amino acids 102 -184), and five-alpha-helical (amino acids 201–303). The region of five different alpha-helical plays a crucial role in dimerizing the 3CL protease since monomeric units lack catalytic activity [37, 38]. At the same time, the series of the amino acid residues such as Glu166, Lue141, Asn142, Cys145, His41, Ser46, Gln187, Thr190, Ala191 and, Pro168 were the catalytic cavity sites [38]. Thus, the binding interaction of the brown algal phlorotannin with 3CL protease displays commonly at the region of catalytic cavity sites. Besides, none of the compounds had exhibited binding interactions with the alpha-helical region of 3CL protease. However, the strong interaction of the compounds at the core region of the catalytic cavity leads to adverse loss of activity. The output of the docking results with the 3CL protease disclosed that the amino acid interaction predominantly occurred at Glu166 in every phlorotannin compound, as shown in Table 2. Secondly, His41, Cys145, and, Asn142 amino acid interactions with phlorotannin compounds were highly observed in the docking pose. Previously, the essentiality of the His41 and Cys145 was noticed [38]. Wherein the catalytic sites of His41 and Cys145 were involved in the proteolytic mechanism of the main and side chain of 3CL proteases. So based on the previous literature, the binding interactions of brown algal phlorotannins were primarily observed in the binding pockets and active sites of 3CLpro, which could potentially intrude the catalytic activity of 3CL protease.
Molecular dynamics (MD) simulation
To further confirm the molecular docking insights, the 200 ns MD simulations were conducted for the SARS-CoV-2 3CLpro with top-ranked marine brown algal phlorotannins (difucol hexaacetate, diphlorethol pentaacetate, eckol hexaacetate, fucofuroeckol) to assess the structural stability and dynamics of the complex molecule of protein–ligand interaction by Schrödinger’s Maestro platform. The complex molecule stability is discussed based on root mean square deviation (RMSD) graph and the protein–ligand contact was elaborated based on the hydrogen bonds, hydrophobic, ionic and water bridges of the total simulation time. The RMSD value of the complex molecule, state of stability with 1000 projection of 200 ns simulation time, is depicted in Fig. 3. The peak values ranged below or around 3 Å distance, representing good stability across the simulation time. The RMSD value of the complex molecule is considerable when the range is around 3 Å distance, which follows in our study [40].
RMSD calculations
The RMSD of 3CLpro and difucol hexaacetate complex have shown stability in their interaction, where the deviations were observed in three phases (Fig. 3a). The minimal peak fluctuations represent the stability of the complex molecule (Fig. 3). The first phase was occupied in the range of 2 Å distance with 137 trajectory projectiles, slightly deviating to 3 Å RMSD to enter the second phase with a deviation of 1 Å distance from the initial phase. The second phase lasted long for up to 919 trajectory projectiles, and considerably a strong interaction was attained between the complex molecules representing the stable nature. In the final phase, the hike of peaks represents the escape of small molecule from the cavity of the complex molecules. The small molecule deviated from the cavity with a distance of 2 Å from the previous phase, and overall, RMSD remained between 3 and 5 Å.
Similarly, the RMSD of 3CLpro and diphlorethol pentaacetate complex molecules showed stability in their interactions (Fig. 3b). The respective RMSD peaks represent three-phase shifts; the first phase layer surrounds 2–3 Å between 200 and 300 trajectories. The second phase entered with a deviation of 4 Å RMSD, and stability was retained in the final phase with the RMSD of 3.5 Å. Thus, complex molecules were stabilized by holding long-time interactions with observable deviations observed in the second phase implying small molecules’ escape from the cavity.
Likewise, the RMSD of 3CLpro with fucofuroeckol complex molecule had shown stability in their interaction. In this complex, the first phase of RMSD showed minimal deviation across 2 Å with nearly 316 trajectories, and the second phase showed as 3.5 Å RMSD with a deviation of 1 Å distance from the initial phase (Fig. 3c). Thus, stability was retained in the range of 2.5 Å distance, representing more stability throughout the total simulation time.
Moreover, the RMSD of the 3CLpro-eckol hexaacetate complex was observed in two phases (Fig. 3d). The first phase remained more stable around the RMSD of 2 Å with 630 trajectories. However, further major deviations were observed representing the ligands’ escape from the scaffold region by deviation up to 6 to 8 Å distance in 981 trajectories that appear out of pocket but still have notable interactions.
The Root Mean Square Fluctuation (RMSF) value of the macromolecule and the small molecule is illustrated in Figs. 4 and 5. In addition, the interactions of the complex molecule across the simulation are shown, such as protein contact timeline, protein secondary structure elements (SSE) like alpha-helices and β-strands, which were monitored throughout the total simulation time (Supplementary Fig. 2).
The stability of complex molecules is also determined by the protein–ligand contact (Fig. 6), in which the interaction should be more than 30% of the total simulation time. Where 0.1 represents 10% and values greater than 0.3 are considered as stable complex molecule. Stability of the complex molecule is based on the hydrogen bonds, water bridges, ionic, and hydrophobic interactions (Schrödinger Release 2021–3). In this study, difucol hexaacetate showed polar interaction with Thr26 for 67% of the total simulation time. Similarly, diphlorethol pentaacetate showed hydrogen bondings with Gln189, Thr190, and Glu166 for 56, 36, and 70%, of total simulation time, respectively. Further, eckol hexaacetate attributed hydrophobic and water bridge interactions at Leu27 (45%) and Glu166 (70%). Furthermore, fucofuroeckol was showed polar interaction with Thr190, Glu166, Gln192 for 53, 70, 65%, and water bridges interaction in Gln189 for 55% of total simulation time.
The total trajectory time was set to 200 ns, where difucol hexaacetate initiated the interaction with Gly143, Glu166, Gln189 and further shifted to continuous contact with Thr26, Asn142. Likewise, diphlorethol pentaacetate initiated the interaction with Gly143, Ser144, Cys145, and shifted to continuous contact with His41, Glu166, Gln189. Besides, eckol hexacetate started interactions with His41, Cys145, and interactions shifted contact to Glu166 and further shifted to Arg298. Also, fucofuroeckol showed continuous interaction with Glu166, His41 and shifted further to Thr190, Gln192, Asn142.
Based on these MD simulation results, Glu166, Gln189, Cys145, and Thr190 tetrad showed the possible interaction site to inhibit the potential activity of 3CLpro. Thus, these results are indicated that phlorotannin compounds could be an effective antagonist to the novel SARS-CoV-2, showing high interaction with the specific targets. However, further investigations are needed to prove its efficacy as an antiviral agent, especially against SARS-CoV-2 using in vitro studies.
Pharmacokinetics and metabolic description of antiviral ligands for drug-likeness assessment
Developing an efficient drug with incompetence in the aspect of ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) is plainly considered as impotent and failure. Firstly, the drug's morphological and metabolic patterns, such as molecular weight, lipophilicity, donor and acceptor hydrogen, human oral adsorption, rules of five, and rules of three, were predominantly evaluated to access the drug-likeness of the phlorotannin ligand molecules. In this study, Jorgensen’s rule of three and Lipinski violations (rules of three) were observed with greater and moderate ranks, which revealed the potential druglike nature of ligands except for dioxinodehydroeckol and diphlorethol. In contrast, the eckol hexacetate, diphlorethol pentaacetate, and difucol hexacetate displayed a good ranking in Lipinski violations but lacked Jorgensen’s rule of three, as shown in Table 5. These observations have clearly comprehended the bioavailability of the phlorotannin ligand molecules as an oral drug candidate. Secondly, followed by the first-pass metabolism, the adsorption, polarity, permeability, and solubility of the compounds are important parameters to be analyzed to outline the proper distribution and metabolism. Especially the computationally predicted human oral adsorption, QPpolrz, (polarity), QplogS (aqueous solubility) and QPPCaco (permeability of the gut-blood barrier) appeared at an adequate level as compared with standard drugs. However, the percentage of human oral adsorption was indirectly proportional to Jorgensen’s rule of three and Lipinski's violations. Furthermore, the #metab (metabolic reaction), QPlogBB (blood–brain barrier) were also observed under limitation as per the standard volume, which can be seen in moderate levels in all of the ligands except phlorofucofuroeckol A and B, fucodiphloroethol G, tetrafucol A, tetraphlorethol, fucotriphlorethol, diphlorethohydroxycarmalol.
Furthermore, the determination of the administration route was comparatively investigated by analyzing the results obtained from the standard drug used against COVID-19, such as remdesivir, ritonavir, favipiravir, lopinavir and hydroxychloroquine. The human oral adsorption, MDCK cell permeability, were considered with standard drug wherein, the hydroxychloroquine exhibited higher human oral adsorption around 91% whereas, the most employed remdesivir effectively used against COVID-19 showed only 36% of oral adsorption [40]. To our surprise, the dioxinodehydroeckol, diphlorethol, diphlorethol pentaacetate, and difucol hexacetate manifested a higher degree of oral absorption (58.0, 58.05, 58.84, 47.30, respectively) as compared with standard remdesivir. Likewise, diphlorethol pentaacetate is the only compound that exhibited a higher value within the given range (25 to 500), about 70.56 in the MDCK permeability. However, it's noteworthy to mention the MDCK value obtained by standard remdesivir was just 6, examined by da Silva et al. [39]. In contrast, the lopinavir revealed higher MDCK permeability with a score of 678 (higher than the limitation described by the QikProp manual), leading to an elevated level of non-active transport in the cells.
Finally, based on all possible parameters and the comparative investigation with standard drugs, the bifuhalol, 2-Phloroeckol, triphloroethol, fucofuroeckol B, fucophlorethol A, diphlorethol pentaacetate, difucol hexacetate, appeared to be the promising candidate with drug-likeness feature predicted by Schrödinger’s QikProp manual.
Conclusion
Identifying alternative and effective therapeutic candidates from biological sources could be a sustainable way to reinforce the drug discovery process against COVID-19. Marine macroalgae are underexploited and an excellent source of bioactive compounds like phlorotannins with proven antiviral and immunostimulatory properties. In this study, phlorotannins were computationally investigated for antagonist effects on multiple protein targets of SARS-CoV-2 (3CLpro, Spro, and RdRp). Interestingly, among the twenty compounds examined, eckol hexacetate, phlorofucofuroeckol B, fucofuroeckol, bifuhalol hexacetate and phlorofucofuroeckol A had shown very efficient inhibition with all the three target proteins with significant binding affinities. Further it was also found that the top ligand molecules against 3CLpro, RdRp, and Spro were eckol hexaacetate, dioxinodehydroeckol, and bifuhalol hexaacetate, respectively, and the phlorotannins were validated with MD simulations, and the distribution of Glu166, Gln189, Cys145, and Thr190 tetrad highlighted the potential interaction sites to inhibit the activity of 3CLpro. Moreover, the phlorotannins were analyzed to be druggable with no major violations from all the ADMET profiling parameters. Therefore, these phlorotannins could be bioactive candidates against SARS-CoV-2 as they can inhibit multiple protein targets activity. However, further investigations are needed to be proven for its efficacy as an antiviral agent, especially against SARS-CoV-2 using in vitro studies. Conclusively, the novel findings of this study could notably impact the advancement of antiviral agent’s interventions for COVID-19 in near future.
Abbreviations
- ADT:
-
AutoDockTools
- PDB:
-
Protein Data Bank
- RMSF:
-
Root mean square fluctuation
- RMSD:
-
Root mean square distance
- 3CLpro:
-
3CL protease
- RdRp:
-
RNA-dependent RNA polymerase
- Spro:
-
Spike glycoprotein
- SARS-CoV-1:
-
Severe acute respiratory syndrome coronavirus
- COVID-19:
-
Coronavirus Disease 2019
- SARS-CoV-2:
-
Novel coronavirus 2019 or 2019-nCov
- SDF:
-
Structure Data File
- MD:
-
Molecular dynamics
- ADMET:
-
Absorption, distribution, metabolism, excretion and toxicity
- MIV-150:
-
MIV-150/Zinc acetate carrageenan
References
Li F (2016) Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3(1):237–261. https://doi.org/10.1146/annurev-virology-110615-042301
Zhou P, Yang XL, Wang XG et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. https://doi.org/10.1038/s41586-020-2012-7
Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R (2020) A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis. Proofread Final Capping Cells 9(5):1267. https://doi.org/10.3390/cells9051267
Hulswit RJ, de Haan CA, Bosch BJ (2016) Coronavirus spike protein and tropism changes. Adv Virus Res 96:29–57. https://doi.org/10.1016/bs.aivir.2016.08.004
Rathnayake AD, Zheng J, Kim Y, Perera KD, Mackin S, Meyerholz DK, Kashipathy MM, Battaile KP, Lovell S, Perlman S, Groutas WC, Chang KO (2020) 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV infected mice. Sci Transl Med 12(557):eabc5332. https://doi.org/10.1126/scitranslmed.abc5332
Jin Z, Wang H, Duan Y, Yang H (2020) The main protease and RNA-dependent RNA polymerase are two prime targets for SARS-CoV-2. Biochem Biophys Res Commun 538:63–71. https://doi.org/10.1016/j.bbrc.2020.10.091
Harvey WT, Carabelli AM, Jackson B, Gupta RK et al (2021) SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19(7):409–424. https://doi.org/10.1038/s41579-021-00573-0
Huang Y, Yang C, Xu XF, Xu W, Liu SW (2020) Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin 41(9):1141–1149. https://doi.org/10.1038/s41401-020-0485-4
Kumar A, Loharch S, Kumar S, Ringe RP, Parkesh R (2020) Exploiting cheminformatic and machine learning to navigate the available chemical space of potential small molecule inhibitors of SARS-CoV-2. Comput Struct Biotechnol J 19:424–438. https://doi.org/10.1016/j.csbj.2020.12.028
Alam MA, Parra-Saldivar R, Bilal M, Afroze CA, Ahmed MN, Iqbal HMN, Xu J (2021) Algae-derived bioactive molecules for the potential treatment of SARS-CoV-2. Molecules 26(8):2134. https://doi.org/10.3390/molecules26082134
Yim SK, Kim K, Kim IH, Chun SH, Oh TH, Kim JU, Kim JW, Jung WH, Moon HS, Ku BS, Jung KJ (2021) Inhibition of SARS-CoV-2 virus entry by the crude polysaccharides of seaweeds and abalone viscera in vitro. Mar Drugs 19(4):219. https://doi.org/10.3390/md19040219
Li YX, Wijesekara I, Li Y, Kim SK (2011) Phlorotannins as bioactive agents from brown algae. Process Biochem 46(12):2219–2224. https://doi.org/10.1016/j.procbio.2011.09.015
Venkatesan J, Keekan KK, Anil S, Bhatnagar I, Kim SK (2019) Phlorotannins. Encyclopedia Food Chem. https://doi.org/10.1016/B978-0-08-100596-5.22360-3
Holdt SL, Kraan S (2011) Bioactive compounds in seaweed: functional food applications and legislation. J Appl Phycol 23(3):543–597. https://doi.org/10.1007/s10811-010-9632-5
Park JY, Kim JH, Kwon JM, Kwon HJ, Jeong HJ, Kim YM, Kim D, Lee WS, Ryu YB (2013) Dieckol, a SARS-CoV 3CL(pro) inhibitor, isolated from the edible brown algae Ecklonia cava. Bioorg Med Chem 21(13):3730–3737. https://doi.org/10.1016/j.bmc.2013.04.026
Cho HM, Doan TP, Ha TKQ, Kim HW, Lee BW, Pham HTT, Cho TO, Oh WK (2019) Dereplication by high-performance liquid chromatography (HPLC) with quadrupole-time-of-flight mass spectroscopy (qTOF-MS) and antiviral activities of phlorotannins from ecklonia cava. Mar Drugs 17(3):149. https://doi.org/10.3390/md17030149
Kim EB, Kwak JH (2015) Antiviral phlorotannin from Eisenia bicyclis against human papilloma virus in vitro. Planta Med 81(16):22. https://doi.org/10.1055/s-0035-1565646
Artan M, Li Y, Karadeniz F, Lee SH, Kim MM, Kim SK (2008) Anti-HIV-1 activity of phloroglucinol derivative, 6,6’-bieckol, from Ecklonia cava. Bioorg Med Chem 216(17):7921–7926. https://doi.org/10.1016/j.bmc.2008.07.078
Kwon HJ, Ryu YB, Kim YM, Song N, Kim CY, Rho MC, Jeong JH, Cho KO, Lee WS, Park SJ (2013) In vitro antiviral activity of phlorotannins isolated from Ecklonia cava against porcine epidemic diarrhea coronavirus infection and hemagglutination. Bioorg Med Chem 21(15):4706–4713. https://doi.org/10.1016/j.bmc.2013.04.085
Yang HK, Jung MH, Avunje S, Nikapitiya C, Kang SY, Ryu YB, Lee WS, Jung SJ (2018) Efficacy of algal Ecklonia cava extract against viral hemorrhagic septicemia virus (VHSV). Fish Shellfish Immunol 72:273–281. https://doi.org/10.1016/j.fsi.2017.10.044
Eom SH, Moon SY, Lee DS, Kim HJ, Park K, Lee EW, Kim YM (2015) In vitro antiviral activity of dieckol and phlorofucofuroeckol-A isolated from edible brown alga Eisenia bicyclis against murine norovirus. Algae 30(3):241–246. https://doi.org/10.4490/algae.2015.30.3.241
Shrestha S, Zhang W, Smid SD (2020) Phlorotannins: a review on biosynthesis, chemistry and bioactivity. Food Biosci 39:100832. https://doi.org/10.1016/j.fbio.2020.100832
Arunkumar M, Gunaseelan S, KubendranAravind M, Mohankumar V, Anupam P, Harikrishnan M, Siva A, Ashokkumar B, Varalakshmi P (2021) Marine algal antagonists targeting 3CL protease and spike glycoprotein of SARS-CoV-2: a computational approach for anti-COVID-19 drug discovery. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.1921032
Besednova NN, Andryukov BG, Zaporozhets TS, Kryzhanovsky SP, Fedyanina LN, Kuznetsova TA, Zvyagintseva TN, Shchelkanov MY (2021) Antiviral effects of polyphenols from marine algae. Biomedicines 9(2):200. https://doi.org/10.3390/biomedicines9020200
Lopez-Santamarina A, Miranda JM, Mondragon ADC, Lamas A, Cardelle-Cobas A, Franco CM, Cepeda A (2020) Potential use of marine seaweeds as prebiotics: a review. Molecules 25(4):1004. https://doi.org/10.3390/molecules25041004
Piccolella S, Crescente G, Faramarzi S, Formato M, Pecoraro MT, Pacifico S (2020) Polyphenols versus coronaviruses: how far has research moved forward? Molecules 25(18):4103. https://doi.org/10.3390/molecules25184103
Al-Khafaji K, Saygili EI, Taskin-Tok T, Cetin Z, Sayın S, Ugur S, Karaaslan MG, Aktas OC, Khan H (2021) Investigation of promising antiviral candidate molecules based on algal phlorotannin for the prevention of COVID-19 pandemic by in silico studies. Biochem Mol Biol 7(1):3
Alexpandi R, De Mesquita JF, Pandian SK, Ravi AV (2020) Quinolines-based SARS-CoV-2 3CLpro and RdRp inhibitors and Spike-RBD-ACE2 inhibitor for drug-repurposing against COVID-19: an in silico analysis. Front Microbiol 11:1796. https://doi.org/10.3389/fmicb.2020.01796
Kandeel M, Al-Nazawi M (2020) Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci 251:117627. https://doi.org/10.1016/j.lfs.2020.117627
Karadeniz FK, Park JW, Park SJ, Kim SK, Egan WJ (2014) Anti-HIV1 activity of phlorotannin derivative 8,4-dieckol from Korean brown alga Ecklonia cava. Biosci Biotechnol Biochem 78:1151–1158. https://doi.org/10.1080/09168451.2014.923282
Aatif M, Muteeb G, Alsultan A, Alshoaibi A, Khelif BY (2021) Dieckol and its derivatives as potential inhibitors of SARS-CoV-2 spike protein (UK Strain: VUI 202012/01): a computational study. Mar Drugs 19(5):242. https://doi.org/10.3390/md19050242
Adegboyega AE, Johnson TO, Omale S (2021) Computational modeling of the pharmacological actions of some antiviral agents against SARS-CoV-2. Data Science for COVID-19 (pp 467–482) Doi: https://doi.org/10.1016/B978-0-12-824536-1.00018-6
Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L (2020) Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368(6492):779–782. https://doi.org/10.1126/science.abb7498
Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P (2020) Structure of replicating SARS-CoV-2 polymerase. Nature 584(7819):154–156. https://doi.org/10.1038/s41586-020-2368-8
Kirchdoerfer RN, Ward AB (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10(1):1–9. https://doi.org/10.1038/s41467-019-10280-3
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807):215–220. https://doi.org/10.1038/s41586-020-2180-5
Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R (2002) Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J 21(13):3213–3224. https://doi.org/10.1093/emboj/cdf327
Kneller DW, Phillips G, O’Neill HM, Jedrzejczak R, Stols L, Langan P, Kovalevsky A (2020) Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat Commun 11(1):1–6. https://doi.org/10.1038/s41467-020-16954-7
Hage-Melim L, Federico LB, de Oliveira N, Francisco V, Correia LC, de Lima HB, Gomes SQ, Barcelos MP, Francischini I, da Silva C (2020) Virtual screening, ADME/Tox predictions and the drug repurposing concept for future use of old drugs against the COVID-19. Life Sci 256:117963. https://doi.org/10.1016/j.lfs.2020.117963
Schrödinger Release 2021–3: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2021 Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2021
Ibrahim MA, Abdelrahman AH, Mohamed TA et al (2021) In Silico mining of terpenes from red-sea invertebrates for SARS-CoV-2 main protease (Mpro) inhibitors. Molecules 26(7):2082. https://doi.org/10.3390/molecules26072082
Ibrahim MA, Abdelrahman AH, Atia MA, Mohamed TA, Moustafa MF, Hakami AR, Khalifa SA, Alhumaydhi FA, Alrumaihi F, Abidi SH, Allemailem KS (2021) Blue biotechnology: computational screening of sarcophyton cembranoid diterpenes for SARS-CoV-2 main protease inhibition. Mar Drugs 19(7):391. https://doi.org/10.3390/md19070391
Martins A, Vieira H, Gaspar H, Santos S (2014) Marketed marine natural products in the pharmaceutical and cosmeceutical industries: tips for success. Mar Drugs 12(2):1066–1101. https://doi.org/10.3390/md12021066
Gentile D, Patamia V, Scala A, Sciortino MT, Piperno A, Rescifina A (2020) Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: a virtual screening and molecular modeling study. Mar Drugs 18(4):225. https://doi.org/10.3390/md18040225
Acknowledgements
The authors thank the Department of Biotechnology (DBT), New Delhi, India (No: BT/PR15677/AAQ/3/799/2016), for the financial support to PV, and International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi for molecular dynamic simulations & ADMET studies using Schrodinger software. The authors also acknowledge DST-PURSE, UGC-UPE, UGC-SAP, RUSA-MKU and DST-FIST facility of Madurai Kamaraj University, Madurai.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
SG: Conceptualization, Validation, Writing-Reviewing and Editing. MA: Methodology, Software, Visualization. MKA: Formal analysis, Software, Writing—Original Draft. SG: Writing-Draft preparation, Reviewing. SR: Software, Resources. VM: Software, Resources. BA: Resources, Supervision. PV: Conceptualization, Project administration, Supervision, Visualization, Validation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sathaiah Gunaseelan, Malaisamy Arunkumar, Manikka Kubendran Aravind these authors have contributed equally to this work
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gunaseelan, S., Arunkumar, M., Aravind, M.K. et al. Probing marine brown macroalgal phlorotannins as antiviral candidate against SARS-CoV-2: molecular docking and dynamics simulation approach. Mol Divers 26, 3205–3224 (2022). https://doi.org/10.1007/s11030-022-10383-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10383-y