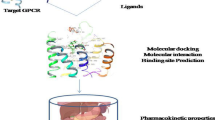
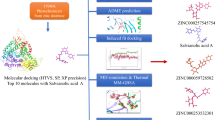
Abstract
The American Cancer Society claims that breast cancer is the second most significant cause of cancer-related death, with over one million women diagnosed each year. Breast cancer linked to the BRCA1 gene has a significant risk of mortality and recurrence and is susceptible to alteration or over-expression, which can lead to hereditary breast cancer. Given the shortage of effective and possibly curative treatments for breast cancer, the present study combined molecular and computational analysis to find prospective phytochemical substances that can suppress the mutant gene (BRCA1) that causes the disease. Virtual screening and Molecular docking approaches are utilized to find probable phytochemicals from the ZINC database. The 3D structure of mutant BRCA1 protein with the id 3PXB was extracted from the NCBI-PDB. Top 10 phytochemical compounds shortlisted based on molecular docking score between − 11.6 and − 13.0. Following the ADMET properties, only three (ZINC000085490903 = − 12.50, ZINC000085490832 = − 12.44, and ZINC000070454071 = − 11.681) of the 10 selected compounds have drug-like properties. The molecular dynamic simulation study of the top three potential phytochemicals showed stabilized RMSD and RMSF values as compared to the APO form of the BRCA1 receptor. Further, trajectory analysis revealed that approximately similar radius of gyration score tends to the compactness of complex structure, and principal component and cross-correlation analysis suggest that the residues move in a strong correlation. Thermostability of the target complex (B-factor) provides information on the stable energy minimized structure. The findings suggest that the top three ligands show potential as breast cancer inhibitors.
Graphical abstract











Similar content being viewed by others
Data availability
Data will be made available on reasonable request.
References
Feng Y, Spezia M, Huang S et al (2018) Breast cancer development and progression: risk factors, cancer stem cells, signalling pathways, genomics, and molecular pathogenesis. Genes Dis 5(2):77–106
Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK (2010) Various types and management of breast cancer: an overview. J Adv Pharm Technol Res 1(2):109–126
Petrucelli N, Daly MB, Feldman GL (2010) Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet Med 12(5):245–259
Deng CX, Scott F (2000) Role of the tumour suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene 19(8):1059–1064
Huang RX, Zhou PK (2020) DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct Target Ther 5(1):60
Roy R, Chun J, Powell SN (2011) BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12(1):68–78
Prakash R, Zhang Y, Feng W, Jasin M (2015) Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 7(4):a016600
Deng CX (2006) BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 34(5):1416–1426
Zheng L, Li S, Boyer TG, Lee WH (2000) Lessons learned from BRCA1 and BRCA2. Oncogene 19(53):6159–6175
Gudmundsdottir K, Ashworth A (2006) The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 25(43):5864–5874
Mehrgou A, Akouchekian M (2016) The importance of BRCA1 and BRCA2 genes mutations in breast cancer development. Med J Islam Repub Iran 30:369
Godet I, Gilkes DM (2017) BRCA1 and BRCA2 mutations and treatment strategies for breast cancer. Integr Cancer Sci Ther. https://doi.org/10.15761/ICST.1000228
Mersch J, Jackson MA, Park M, Nebgen D, Peterson SK, Singletary C, Arun BK, Litton JK (2015) Erratum: cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 121(14):2474–2475
Gaonkar R, Avti PK, Hegde G (2018) Differential antifungal efficiency of geraniol and citral. Nat Prod Commun. https://doi.org/10.1177/1934578X1801301210
Pinzi L, Rastelli G (2019) Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci 20(18):4331
Meng XY, Zhang HX, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7(2):146–157
Zhou HX, Pang X (2018) Electrostatic interactions in protein structure, folding, binding, and condensation. Chem Rev 118(4):1691–1741
Zhang S (2011) Computer-aided drug discovery and development. Methods Mol Biol 716:23–38
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48(3):443–453
Zhang Z, Li Y, Lin B, Schroeder M, Huang B (2011) Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics 27(15):2083–2088
Elrod P, Zhang J, Yang X et al (2002) Contributions of active site residues to the partial and overall catalytic activities of human S-adenosylhomocysteine hydrolase. Biochemistry 41(25):8134–8142
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
Banerjee P, Eckert AO, Schrey AK, Preissner R (2018) ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res 46(W1):W257–W263
Couch FJ, Weber BL (1996) Mutations and polymorphisms in the familial early-onset breast cancer (BRCA1) gene. Breast Cancer Inf Core Hum Mutat 8(1):8–18
Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmañà J et al (2012) Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov 2(11):1048–1063
Rehman FL, Lord CJ, Ashworth A (2012) The promise of combining inhibition of PI3K and PARP as cancer therapy. Cancer Discov 2(11):982–984
Martorana F, Motta G, Pavone G, Motta L, Stella S, Vitale SR, Manzella L, Vigneri P (2021) AKT inhibitors: new weapons in the fight against breast cancer? Front Pharmacol 29(12):546
Aziz D, Portman N, Fernandez KJ, Lee C, Alexandrou S, Llop-Guevara A, Phan Z, Yong A, Wilkinson A, Sergio CM, Ferraro D (2021) Synergistic targeting of BRCA1 mutated breast cancers with PARP and CDK2 inhibition. NPJ Breast Cancer 7(1):1–4
Choudhari AS, Mandave PC, Deshpande M, Ranjekar P, Prakash O (2020) Phytochemicals in cancer treatment: from preclinical studies to clinical practice. Front Pharmacol 28(10):1614
Khanduja KL, Kumar S, Varma N, Varma SC, Avti PK, Pathak CM (2008) Enhancement in alpha-tocopherol succinate-induced apoptosis by all-trans-retinoic acid in primary leukemic cells: role of antioxidant defense, Bax and c-myc. Mol Cell Biochem 319(1–2):133–139
Kumar A, Nisha CM, Silakari C et al (2016) Current and novel therapeutic molecules and targets in Alzheimer’s disease. J Formos Med Assoc 115(1):3–10
Jayaraman S, Veeraraghavan V, Sreekandan RN, Mohan SK, Suga S, Kamaraj D, Mohandoss S, Koora S (2020) Molecular docking analysis of the BRCA1 protein with compounds from Justica adhatoda L. Bioinformation 16(11):888–892
Prabhavathi H, Dasegowda KR, Renukananda KH, Lingaraju K, Naika HR (2021) Exploration and evaluation of bioactive phytocompounds against BRCA proteins by in silico approach. J Biomol Struct Dyn 39(15):5471–5485
Kumar S, Khanduja KL, Verma N, Verma SC, Avti PK, Pathak CM (2008) ATRA promotes alpha tocopherol succinate-induced apoptosis in freshly isolated leukemic cells from chronic myeloid leukemic patients. Mol Cell Biochem 307(1–2):109–119
Coquelle N, Green R, Glover JN (2011) Impact of BRCA1 BRCT domain missense substitutions on phosphopeptide recognition. Biochemistry 50(21):4579–4589
Souza PCT, Thallmair S, Conflitti P et al (2020) Protein-ligand binding with the coarse-grained martini model. Nat Commun 11(1):3714
Lippert T, Rarey M (2009) Fast automated placement of polar hydrogen atoms in protein-ligand complexes. J Cheminform 1(1):1–2
Shih AJ, Telesco SE, Choi SH, Lemmon MA, Radhakrishnan R (2011) Molecular dynamics analysis of conserved hydrophobic and hydrophilic bond-interaction networks in ErbB family kinases. Biochem J 436(2):241–251
Yu W, MacKerell AD Jr (2017) Computer-aided drug design methods. Methods Mol Biol 1520:85–106
Kuhlman B, Bradley P (2019) Advances in protein structure prediction and design. Nat Rev Mol Cell Biol 20(11):681–697
Haftarah A, Wang J (1864) Miao Y (2020) Retrospective ensemble docking of allosteric modulators in an adenosine G-protein-coupled receptor. Biochim Biophys Acta Gen Subj 8:129615
Fataftah H, Karain W (2014) Detecting protein atom correlations using correlation of probability of recurrence. Proteins 82(9):2180–2189
Funding
One of the authors, Jitender Singh much acknowledges the Indian Council of Medical Research New Delhi, for awarding ICMR˗SRF (Ref. No.: BMI/11(54)/2020, Dated 24/03/2021) for the financial assistance provided.
Author information
Authors and Affiliations
Contributions
JS performed the study, analyzed and written the manuscript, NS and AC prepared the figures and written the manuscript, PS, AP and BM edited and revised the manuscript PKA Conceptualized, Design, interpretation and written the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflict of Interest.
Ethical approval
This is an in silico study and does not require ethical approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, J., Sangwan, N., Chauhan, A. et al. Screening and identification of phytochemical drug molecules against mutant BRCA1 receptor of breast cancer using computational approaches. Mol Cell Biochem 477, 885–896 (2022). https://doi.org/10.1007/s11010-021-04338-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04338-4