Abstract
Acinetobacter baumannii is one of the most problematic pathogens in clinical settings. Emerging of its antibiotic-resistant strains persuade researchers to find alternative treatment options such as immunization against the notorious nosocomial pathogen. Oma87 has been introduced as an immunogenic outer membrane protein via reverse vaccinology. However, protectivity of A. baumannii Oma87 is not well known. The current research undertakes a study on the immunogenicity of recombinant Oma87 in a murine model. Some physico-chemical properties were assessed via in silico analyses. The corresponding gene was amplified and cloned into pET28a plasmid. The recombinant protein was purified and then was administered to immunize mice. Sera obtained from the immunized mice were assessed with respect to the triggered antibodies. Challenges were performed on actively or passively immunized mice. In silico analyses revealed that this protein is the same as BamA. A high titer of specific antibody was raised against rOma87 even after the first injection. The specific antibody recognized the whole cell of A. baumannii. Both active and passive immunizations confer 100 and 50% protection, respectively against ~ 2 × lethal dose (LD) of A. baumannii in the murine sepsis model. Although none of mice received ~ 5 × LD of A. baumannii survived in passive immunization, 25% of mice challenged with ~ 7 × LD of the bacteria survived and the dead mice exhibited a delayed death. Based on these results, Oma87 is the same as BamA which could be considered as a promising vaccine candidate against A. baumannii in the sepsis model.
Similar content being viewed by others
Introduction
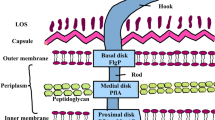
Acinetobacter baumannii is a notorious Gram-negative bacterium assigned as one of the most dangerous nosocomial pathogens (Huang et al. 2016). This serious health threat could cause multiple infection types of meninges, soft tissues, pulmonary, urinary and systemic tissues (McConnell et al. 2013) among which pneumonia is the main one (Fajardo Bonin et al. 2014). The mortality rate of A. baumannii-associated infections is as high as 70% and 60% for pneumonia and bacteremia, respectively (Fajardo Bonin et al. 2014). Recent reports on the emergence of antibiotic-resistant strains could be considered as concerning alarm for the bacterium treatment (Pachón and McConnell 2014). Since there is no commercially effective antibiotic against its rapid emerging multidrug-resistant (MDR) and pan-drug resistant (PDR) strains of the pathogen, clinical management of these successful pathogens is difficult (Pachón and McConnell 2014). Hence, researchers are motivated to benefit from alternative approaches such as active or passive immunization to combat with A. baumannii. For as much as this superbug is an opportunistic pathogen, it is known as a troublesome bacterium in immunocompromised patients (Ahmad et al. 2016). In the aforementioned case, the failure of active immunization is probable (Pachón and McConnell 2014; Perez and Bonomo 2014; Ahmad et al. 2016) highlighting passive immunization via antibodies elicited against engaged antigens. In this respect, the antigens producing protective active and passive immunization could be assumed as appropriate. To date, a number of in silico, in vitro and in vivo investigations have been conducted to nominate or design protein antigens (Fattahian et al. 2011; Bentancor et al. 2012; Luo et al. 2012; Rahbar et al. 2012; Lin et al. 2013; Moriel et al. 2013; Fajardo Bonin et al. 2014; Hosseingholi et al. 2014; Badmasti et al. 2015; Chiang et al. 2015; Huang et al. 2015, 2016 Sefid et al. 2015; Garg et al. 2016; Singh et al. 2016; Jahangiri et al. 2017, 2018a, b Ni et al. 2017; Guo et al. 2018; Song et al. 2018; Bazmara et al. 2017), OmpA (Luo et al. 2012; Lin et al. 2013), OmpW (Huang et al. 2015), Bap (Fattahian et al. 2011), Omp22 (Huang et al. 2016) and FilF (Singh et al. 2016) are among protective antigens against this nosocomial pathogen. Amongst the antigens, outer membrane proteins (OMPs) are considerable owing to their roles in antibiotic resistance, adhesion to host cells and pathogenicity of the bacterium in addition to their high immunogenicity (McConnell et al. 2013; Fajardo Bonin et al. 2014). OMPs are abundant on bacterial cell surface making them suitable target to be engaged as vaccine candidates (Fajardo Bonin et al. 2014). Although some promising immunogens such as OmpA have been remarked (Chen 2015), attempts toward unveiling new targets are in progress. Reverse vaccinology is known as a robust approach to introduce novel targets as appropriate vaccine candidates (Moriel et al. 2013; Chiang et al. 2015; Ni et al. 2017; Ahmad and Azam 2018; Solanki and Tiwari 2018). Oma87 in A. baumannii is one of the immunogenic proteins that has been assigned via reverse vaccinology as an appropriate vaccine candidate (Chiang et al. 2015). Although function of the OMP in A. baumannii as well as its role in pathogenicity of this bacterium remains to be explored, its expression and localization have been confirmed (Mendez et al. 2012; Chiang et al. 2015). In Pasteurella multocida, this protein is nominated as an appropriate immunogen which could confer protection against pasteurellosis in livestock (Kumar et al. 2013). However, no data about protectivity of A. baumannii Oma87 is available. These evidence persuade us to conduct the current study aiming at evaluation of Oma87 immunogenicity in murine model. In the present study, we expressed and purified the recombinant Oma87 of A. baumannii. The purified protein was then administered into mice in order to trigger antibodies. Active and passive immunizations were assessed in a murine model.
Material and Methods
In Silico Analyses
The Oma87 protein sequence with Accession No. WP_002016574.1 was obtained from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/protein) and saved in FASTA format. This protein sequence was defined as the outer membrane protein assembly factor BamA at https://www.ncbi.nlm.nih.gov/protein. BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed using the protein sequence against non-redundant protein database to find similar sequences. Moreover, a BLAST search restricted to Homo sapiens (taxid:9606) was also performed against non-redundant protein database. ProtParam (Gasteiger et al. 2005) (https://web.expasy.org/protparam/) was harnessed to analyze some physicochemical parameters such as molecular weight, amino acid composition, instability index, theoretical pI, aliphatic index, grand average of hydropathicity and half-life of the protein in mammalian, yeast, and Escherichia coli. SignalP 4.1 (Petersen et al. 2011) (https://www.cbs.dtu.dk/services/SignalP) was used to predict the signal peptide and its cleavage site. Toxicity of the protein was predicted by ToxinPred (Gupta et al. 2013) at https://crdd.osdd.net/raghava/toxinpred/. The parameters were set as default.
Bacterial Strains
Acinetobacter baumannii ATCC 19606 and E. coli BL21 (DE3) were used in this study. Acinetobacter baumannii ATCC 19606 was used as a source for oma87 gene. This strain was also employed in whole-cell ELISA as well as mice challenges. Escherichia coli BL21 (DE3) was used to express recombinant Oma87 (rOma87).
rOma87 Production and Purification
Genomic DNA of A. baumannii was extracted and employed as a template for polymerase chain reaction (PCR). oma87 gene was amplified by the primers: ACTGGATCCATGGATGATTTCGTTGTTAGAGATATTC (forward) and GTACCTCGAGTTAGAAAGTACGACCAATTTCAAACTGAAT (reverse). PCR reaction was performed with pre-denaturation at 94 °C for 3 min followed by 35 thermal cycles of denaturation at 94 °C for 40 s, annealing at 62 °C for 45 s, and final extension at 72 °C for 5 min. Amplified PCR products were analyzed by 1% agarose gel electrophoresis and purified with DNA gel extraction kit (Qiagen). The amplified gene was cloned into pET28a(+) between BamHI and XhoI restriction sites and was transformed to competent cells of E. coli BL21 (DE3) prepared by calcium chloride (CaCl2) chemical method. The selected clones were confirmed by both PCR and sequencing. The transformed cells were grown in LB supplemented with 70 μg/ml kanamycin at 37 °C with shaking at 150 rpm. Then a single colony from an agar plate was transferred to 1 ml of ZYP-5052 medium [for 100 ml:93 ml ZY (10 g N-Z-amine AS Sigma cat. # N4S 17, 5 g yeast extract, 925 ml water), 0.1 ml 1 M MgSO4, 2 ml of 50 × 5052 (0.5% glycerol, 0.05% glucose, 0.2% α-lactose), 5 ml 20 × NPS (6.6% of (NH4)2SO4, 13.6% of KH2PO4, 14.2% of Na2HPO4 in double-distilled water), plus antibiotic in a microtube and shake at 37 °C for 6–8 h. Over-expression of the protein was achieved by transferring 1 ml fresh culture of bacteria to 100 ml ZYP-5052 + kanamycin and then was incubated overnight at 37 °C. After incubation, cooling was carried out on the ice buckets after which the cells were harvested by centrifugation at 2630 rcf for 15 min at 4 °C. The cell pellet was resuspended in buffer B (100 mM NaH2PO4, 10 mM Tris–HCl, 8 M urea, pH 8) and then was sonicated (5 times, 15 s, 1 min interval) using a sonicator. The lysate was centrifuged at 17,760 rcf for 20 min at 4 °C to remove the debris. The supernatant was loaded on to the Ni-NTA agarose (Qiagen, Germany) affinity column, pre-equilibrated with the buffer B and cell lysate slowly added (3–4 column volumes per hour) onto the column. The resin was washed twice with washing buffer (100 mM NaH2PO4, 10 mM Tris–HCl, 8 M urea and 20 mM imidazole at pH 8), and the recombinant protein was eluted with 5 times 250 µl elution buffer (100 mM NaH2PO4, 10 mM Tris–HCl, 8 M urea and 250 mM imidazole at pH 8). The elution fractions were monitored by measuring their absorbance at 280 nm. The purified protein was analyzed by 9% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Refolding of the purified recombinant protein was carried out via sequential dialysis against PBS (pH 7.4) containing 6, 4, 2 and 0 M urea + 0.5 mM l-arginine at 4 °C for 2 h. The purified protein concentration was determined with the Bradford method (Bradford 1976) using bovine serum albumin (BSA) as standard. To validate the recombinant protein expression, western blotting with horseradish peroxidase (HRP)-conjugated anti-polyhistidine antibodies (1:10,000 dilution) was performed in which, 0.493 µg of the recombinant protein was loaded onto the SDS-PAGE.
Animal Study and Ethical Clearance
6 to 8 week old healthy female BALB/c mice, weighing 22–25 g were procured from the Baqiyatallah University of Medical Sciences, Tehran, Iran. Six mice were grouped as control and eight mice were used as test groups. The animals were maintained in clean standard conditions in the Animal Care Facility of Shahed University. All the animal tests were performed in accordance with animal care guidelines confirmed by Animal Care and Ethical Committee of Shahed University. The mice were immunized subcutaneously (s.c.) injected in 100 µl volume in Freund’s adjuvant mixture on days 0, 14, 28 and 42. Control mice were immunized with 1:1 (v/v) combination of Freund’s adjuvant (Sigma) and PBS containing 0.5 M arginine. The first injection was carried out with complete Freund’s adjuvant and the subsequent ones were done with incomplete Freund’s adjuvant. For test groups, 20 μg of antigen at 1:1 (v/v) ratio of Freund’s adjuvant and PBS containing 0.5 M arginine, was administered.
Enzyme-Linked Immunosorbent Assay (ELISA)
The IgG titer against the recombinant protein in the sera obtained from the immune mice was assayed by ELISA. Briefly, 100 μl of the purified recombinant Oma87 (20 μg/ml) was coated in a 96-well microtiter plate and the plate was incubated overnight at 4 °C. The wells were washed thrice with PBST and were then blocked with 100 μl of PBST plus 5% skimmed milk. The plate was incubated at 37 °C for 1 h after which was washed three times with PBST. Serial dilutions at 1:250 to 1:64,000 range of mice sera were added to the wells and the plate was incubated at 37 °C for 2 h. The washing step was repeated and then 100 μl of anti-mouse IgG conjugated with HRP at 1:15,000 dilution was added to each well. The plate was incubated at 37 °C for 1 h. After repeating the washing step, 100 μl of TMB (3, 3′, 5, 5′-tetramethylbenzidine) was added to each well. The plate was kept in dark at room temperature for 15 min to develop color change. The reaction was stopped by addition of 100 μl of 3 M H2SO4 per well and the optical density was determined at 450 nm using an ELISA plate reader.
Whole-Cell ELISA
The A. baumannii ATCC 19606 was grown in LB at 37 °C to reach OD600 of 0.6. The bacteria suspended (108 cells/ml) in coating buffer (50 mM sodium carbonate buffer, pH 9.6) were coated (100 μl/well) in the wells overnight at 4 °C. To complete bacterial attachment the plate was dried under warm air blow. The wells were then washed thrice with PBST. PBST plus 5% skimmed milk (100 μl/well) was added as a blocking buffer followed by incubating at 37 °C for 1 h. Serial dilutions (1:100 to 1:3200) of mice sera were added to the wells after washing three times with PBST. The plate was then incubated at 37 °C for 2 h. The washing step was repeated and then 100 μl of anti-mouse IgG conjugated with HRP at 1:15,000 dilution was added to each well. The plate was incubated at 37 °C for 1 h. After repeating washing step, 100 μl TMB was added to each well. The plate was kept in dark at room temperature for 15 min to develop color. The reaction was stopped by adding 3 M H2SO4 (100 μl/well).
Western Blotting with Anti-serum (Immunoblotting)
346 µg of the rOma87 was electrophoresed by a 9% SDS-PAGE gel and then transferred on to a nitrocellulose membrane. The membrane was blocked with 5% skimmed milk in PBST and then incubated with 1:4000 dilution of the sera collected from the immunized mice. HRP-coupled anti-mouse IgG was used as a secondary antibody and then washed with PBST three times. The visualization of membrane was developed by DAB solution. The chromogenic reaction was stopped by PBST.
Challenge of Mice with A. baumannii ATCC 19606
Lethal dose (LD) of A. baumannii ATCC 19606 was determined. The prepared bacteria were mixed with 10% porcine mucin prior to intraperitoneal injection. The immunized mice were intraperitoneally challenged with 2 × 106 CFU or 7 × 106 CFU of A. baumannii ATCC 19606. The survival trend of mice was monitored for 4 days. Passive immunization was carried out as follows: the sera complements were inactivated by incubating at 56 °C for 30 min. 100 μl of non-immune or immune undiluted sera were intravenously injected into each of the control or test groups. After 3 h, the mice received intraperitoneally 2 × 106 CFU or 5 × LD of A. baumannii ATCC 19606. The survival rate of mice was considered for 4 days.
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism 7.03. The data were presented as mean and standard deviations were represented as error bars. One-way ANOVA with post hoc Scheffé tests was employed to analyze the statistical significance of results. Results with p values of < 0.05 were considered as significant.
Results
In Silico Analysis
The Accession No. of WP_002016574.1 covers more than 2400 strains of A. baumannii. BLAST search found 29 hits called BamA, YaeT or membrane protein with an identity of ≥ 99%, query coverage: 100% and E value 0. These hits cover more than 2500 strains of A. baumannii. No hit was found in the BLAST search limited to the H. sapiens (taxid:9606). Oma87 is an 841-amino acid protein with theoretical pI 5.26. Its theoretical molecular weight is 93.77 kDa. This protein has four cysteine residues. Oma87 was classified as s stable protein with instability index of 29.44 (proteins with instability index of < 40 are stable). The aliphatic index and the GRAVY value of Oma87 were calculated as 76.46 and − 0.447, respectively. The estimated half-life of the protein in mammalian, yeast and E. coli is 30 h, > 20 h and > 10 h respectively. Cleavage site of signal peptide was predicted between residues 21 and 22 of the protein sequence. DVRKYCEDNYGF was predicted as toxic region within the Oma87 sequence.
Oma87 Amplification and Expression
The amplified gene was 2526 bp. The fragments were confirmed by restriction enzyme digest analysis. The rOma87 was expressed in E. coli BL21 (DE3). Analysis of the SDS-PAGE revealed a band with an apparent molecular weight of approximately 95 kDa is corresponding to rOma87. According to the production of proteins in inclusion bodies, purification was performed as denaturing condition. The eluted proteins were analyzed by SDS-PAGE and western blotting. Western blotting with anti-His-tag antibody confirmed expression of rOma87 (Fig. 1).
IgG Response to Oma87
ELISA showed high titers of specific IgG against the purified recombinant protein. The antibody increase was significant (p < 0.0001) as compared to the control group where no anti-Oma87 antibody was observed (Fig. 2).
Indirect ELISA against rOma87. Two micrograms of the rOma87 was coated in wells. Serial dilutions (1:250 to 1:64,000) of immunized or non-immunized mice sera were added to the wells. p-value of non-immunized mice sera vs. 1st, 2nd, 3rd, and 4th injections were p < 0.01, < 0.001, ≤ 0.0004 and < 0.001, respectively
Whole-Cell ELISA
In whole-cell ELISA, immune mice sera showed higher absorbance than the control mice sera (p < 0.001). Based on the ELISA result, anti-Oma87 antibodies could recognize the native protein expressed on A. baumannii ATCC 19606 (Fig. 3).
Western Blotting with Anti-serum (Immunoblotting)
The Oma87 immunoreactivity was also analyzed by immunoblotting using obtained anti-sera from immunized mice. The antiserum reacted with the purified rOma87.
Animal Challenge Studies
LD of A. baumannii ATCC 19606 was determined as 1 × 106 CFU. All actively immunized mice that received 2 × 106 CFU (~ 2 × LD) of A. baumannii ATCC 19606, survived while all the control mice died 24 h after challenge. Twenty-five percent of mice challenged with 7 × 106 CFU (7 × LD) of bacteria survived while the remaining 75% exhibited a delayed death (Fig. 4). In passive immunization study, 50% of mice receiving 2 × 106 CFU (~ 2 × LD) of A. baumannii ATCC 19606 survived. None of the mice survived in passive immunization at 5 × LD of the bacterial dose (Fig. 5).
Discussion
Oma87 is nominated as a suitable antigen that could be considered for vaccine development against A. baumannii (Chiang et al. 2015). Based on the name of retrieved protein sequence as well as homology search, we found out that this OMP is BamA. Although its function in A. baumannii remains obscure, this highly conserved OMP has pivotal role in outer membrane protein assembly of Gram-negative bacteria. Moreover, BamA structure in E. coli has already been unveiled (Gessmann et al. 2014). Immunoprotectivity of this highly conserved protein against A. baumannii had been recently demonstrated in a murine pneumonia model (Singh et al. 2017). An aliphatic index range of 42.08–90.68 revealed a thermostable protein (higher the aliphatic index shows higher the thermostability of the protein). Based on the calculated aliphatic index, Oma87 is a thermostable protein. GRAVY of a protein is related to its solubility properties. The calculated GRAVY (negative value) for Oma87 indicates that the protein is non-polar.
In terms of conservancy, BLAST results showed that this protein is highly conserved among the different A. baumannii strains. Its prevalence and conservancy is more than FilF (Singh et al. 2016), OmpW (Huang et al. 2015), Omp34 (Jahangiri et al. 2018a, b) and Omp22 (Huang et al. 2016). Moreover, Oma87 shares no similarity with human proteins which is concordant to results obtained in Singh et al. study (Singh et al. 2017). This result implies that Oma87 is a safe antigen with no autoimmune responses in the host (human).
Since this is an outer membrane protein, hydrophobic stretches within these proteins could result in aggregation and formation of the inclusion body where it is overexpressed. Moreover, the existence of signal peptide could promote this phenomenon. Moreover, this protein has four cysteines could form disulfide bonds and therefore has high hydrophobicity. So, this protein was purified in denaturing conditions. Denaturing conditions could disturb structure of proteins consequently their structural epitopes. Hence, the refolding procedure was conducted; however, aggregates were again formed in dialyzing procedure. Based on literature review, adding l-arginine could be helpful for refolding the recombinant protein aggregates (Tsumoto et al. 2004; Chen et al. 2009). A high titer of IgG raised against rOma87 (BamA) reveals its high antigenicity. This finding was in accordance with the previous study on this protein (Singh et al. 2017). Although the amount of recombinant protein administered in the current study is the same as that used in boosters of previous study, the adjuvant as well as route of injection are different (Singh et al. 2017). Whole-cell ELISA results demonstrated that the antibodies triggered against rOma87 (BamA) could recognize the native protein expressed in A. baumannii. In addition, it is revealed this OMP is expressed under in vitro culture condition and is also exposed to these specific antibodies. In general, whole-cell ELISA shows lower absorbance in comparison to the purified protein antigen. It could be due to lower binding sites, exposed to anti-rOma87 antibodies, on bacterial surfaces. Since this antigen is an OMP, it is expected that all residues are not to be accessible for the antibodies raised against rOma87 (BamA). Moreover, other antigenic targets of bacterial cell surface are also present in whole-cell ELISA. It could be expected that removal of non-epitope regions of this immunogen will increase its antigenicity. Moreover, removal of signal peptide as well as non-exposed regions of this antigen could conduct antibodies triggering toward more efficient specific antibodies. Hence, determination of Oma87 (BamA) structure as well as its loops which are exposed in native form would be valuable. Although 3D structure of BamA had been predicted by Singh et al. (2017), topology prediction along with harnessing an integrative in silico approach could improve quality of the predicted model. Recently, structure of Omp34 (a promising antigen of A. baumannii) has been precisely predicted via harnessing an integrative in silico approach (Jahangiri et al. 2018a, b). Pierce into 3D structure of Oma87 (BamA) could help in assigning appropriate exposed antigenic determinant. Active and passive immunizations have been considered as options to be investigated against A. baumannii (McConnell and Pachón 2010; Luo et al. 2012; Huang et al. 2015, 2016; Singh et al. 2016; Jahangiri et al. 2019). In the previous study conducted by Singh et al. (2017), active and passive immunization with this protein conferred 80% and 60% protection, respectively against intranasal challenge with LD of A. baumannii. In the present study, to avoid large quantity of bacteria in inoculums, porcine mucin was used. A large quantity of bacteria administration could result in a model of intoxication with endotoxins rather than a reliable infection model (Cross et al. 1993; Buras et al. 2005). Mucin could increase the virulence of bacteria in murine models. This may lead to employing lower doses of bacteria to establish murine sepsis model reflecting more accurately human sepsis. Active immunization showed full protection against 2 × LD in the murine sepsis model. Moreover, the active immunization showed higher protection than passive immunization. This is in agreement with previous reports (Huang et al. 2015, 2016). It could be due to the presence of other immune system effectors in active immunization in addition to antibodies. Based on the obtained results and findings of Singh et al. (2017), it seems that the systemic immune response triggered by this antigen is more effective in sepsis model than pneumonia. It could be attributed to role of mucosal immunity had not been considered in the previous investigation. It could be predicted that triggering mucosal immunity could confer higher protection against murine pneumonia caused by A. baumannii. In a recent study on immunogenicity of Omp22, the immunized mice were challenged intraperitoneally with LD of A. baumannii. The survival rate of mice actively immunized with 20 µg of this OMP was less than 50%; while all mice immunized with 50 µg of rOmp22 were survived within the follow-up time. Moreover, anti-rOmp22 IgG titers in mice group received 50 µg of rOmp22 were significantly higher than IgG titers in mice received 20 µg of this OMP. In our study on rOma87, only 20 µg of this protein was administered to be investigated. So, it could be deduced that Oma87 (BamA) is a stronger antigen than Omp22 (Huang et al. 2016). Survival results of active and passive immunization imply that this antigen is expressed in the murine model infection. An appropriate vaccine candidate should be highly expressed in infection situation. Although the whole-cell ELISA and challenge results demonstrate in vitro and in vivo expression of Oma87 (BamA) in murine model, its expression level in human infections needs to be explored. The abundance of the protein on bacterial surface as well as its role in various phases of infection could impact on efficacy of the vaccine candidate.
Based on the study results, Oma87 (BamA) could elicit protective antibodies that nominate it as an appropriate antigen to be considered in the active and passive trials against A. baumannii in the sepsis model. Since this is an OMP, determination of exposed regions for future vaccine design attempts would be useful.
References
Ahmad S, Azam SS (2018) A novel approach of virulome based reverse vaccinology for exploring and validating peptide-based vaccine candidates against the most troublesome nosocomial pathogen: Acinetobacter baumannii. J Mol Graph Model 83:1–11. https://doi.org/10.1016/j.jmgm.2018.04.020
Ahmad TA, Tawfik DM, Sheweita SA, Haroun M, El-Sayed LH (2016) Development of immunization trials against Acinetobacter baumannii. Trials Vaccinol 5:53–60. https://doi.org/10.1016/j.trivac.2016.03.001
Badmasti F, Ajdary S, Bouzari S, Fooladi AAI, Shahcheraghi F, Siadat SD (2015) Immunological evaluation of OMV (PagL)+ Bap (1–487aa) and AbOmpA (8–346aa)+ Bap (1–487aa) as vaccine candidates against Acinetobacter baumannii sepsis infection. Mol Immunol 67:552–558. https://doi.org/10.1016/j.molimm.2015.07.031
Bazmara H, Rasooli I, Jahangiri A, Sefid F, Astaneh SDA, Payandeh Z (2017) Antigenic properties of iron regulated proteins in Acinetobacter baumannii: an in silico approach. Int J Pept Res Ther. https://doi.org/10.1007/s10989-017-9665-6
Bentancor LV, Routray A, Bozkurt-Guzel C, Camacho-Peiro A, Pier GB, Maira-Litrán T (2012) Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect Immun 80:3381–3388. https://doi.org/10.1128/IAI.06096-11
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Buras JA, Holzmann B, Sitkovsky M (2005) Model organisms: animal models of sepsis: setting the stage. Nat Rev Drug Discov 4:854. https://doi.org/10.1038/nrd1854
Chen J, Liu Y, Li X, Wang Y, Ding H, Ma G, Su Z (2009) Cooperative effects of urea and l-arginine on protein refolding. Protein Expr Purif 66:82–90. https://doi.org/10.1016/j.pep.2009.02.004
Chen W (2015) Current advances and challenges in the development of Acinetobacter vaccines. Hum Vaccines Immunother 11:2495–2500. https://doi.org/10.1080/21645515.2015.1052354
Chiang M-H, Sung W-C, Lien S-P, Chen Y-Z, Lo AF-Y, Huang J-H, Kuo S-C, Chong P (2015) Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum Vaccines Immunother 11:1065–1073. https://doi.org/10.1080/21645515.2015.1010910
Cross AS, Opal S, Sadoff J, Gemski P (1993) Choice of bacteria in animal models of sepsis. Infect Immun 61:2741–2747
Fajardo Bonin R, Chapeaurouge A, Perales J, Silva JG, do Nascimento HJ, D’Alincourt Carvalho Assef AP, Senna M, Procópio J (2014) Identification of immunogenic proteins of the bacterium Acinetobacter baumannii using a proteomic approach. PROTEOMICS Clin Appl 8:916–923. https://doi.org/10.1002/prca.201300133
Fattahian Y, Rasooli I, Gargari SLM, Rahbar MR, Astaneh SDA, Amani J (2011) Protection against Acinetobacter baumannii infection via its functional deprivation of biofilm-associated protein (Bap). Microb Pathog 51:402–406. https://doi.org/10.1016/j.micpath.2011.09.004
Garg N, Singh R, Shukla G, Capalash N, Sharma P (2016) Immunoprotective potential of in silico predicted Acinetobacter baumannii outer membrane nuclease, NucAb. Int J Med Microbiol 306:1–9. https://doi.org/10.1016/j.ijmm.2015.10.005
Gasteiger E, Hoogland C, Gattiker A, Duvaud SE, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. Springer. https://doi.org/10.1385/1-59259-890-0:571
Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG (2014) Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc Natl Acad Sci USA 111:5878–5883. https://doi.org/10.1073/pnas.1322473111
Guo SJ, Ren S, Xie YE (2018) Evaluation of the protective efficacy of a fused OmpK/Omp22 protein vaccine candidate against Acinetobacter baumannii infection in mice. Biomed Environ Sci 31:155–158. https://doi.org/10.3967/bes2018.019
Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Open Source Drug Discovery C, Raghava GP (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 8:e73957 https://doi.org/10.1371/journal.pone.0073957
Hosseingholi EZ, Rasooli I, Gargari SLM (2014) In silico analysis of Acinetobacter baumannii phospholipase D as a subunit vaccine candidate. Acta Biotheor 62:455–478. https://doi.org/10.1007/s10441-014-9226-8
Huang W, Wang S, Yao Y, Xia Y, Yang X, Long Q, Sun W, Liu C, Li Y, Ma Y (2015) OmpW is a potential target for eliciting protective immunity against Acinetobacter baumannii infections. Vaccine 33:4479–4485. https://doi.org/10.1016/j.vaccine.2015.07.031
Huang W, Yao Y, Wang S, Xia Y, Yang X, Long Q, Sun W, Liu C, Li Y, Chu X (2016) Immunization with a 22-kDa outer membrane protein elicits protective immunity to multidrug-resistant Acinetobacter baumannii. Sci Rep 6:20724. https://doi.org/10.1038/srep20724
Jahangiri A, Owlia P, Rasooli I, Salimian J, Derakhshanifar E, Naghipour Erami A, Darzi Eslam E, Darvish Alipour Astaneh S (2019) Specific egg yolk antibodies (IgY) confer protection against Acinetobacter baumannii in a murine pneumonia model. J Appl Microbiol 126:624–632. https://doi.org/10.1111/jam.14135
Jahangiri A, Rasooli I, Owlia P, Fooladi AAI, Salimian J (2017) In silico design of an immunogen against Acinetobacter baumannii based on a novel model for native structure of Outer membrane protein A. Microb Pathog 105:201–210. https://doi.org/10.1016/j.micpath.2017.02.028
Jahangiri A, Rasooli I, Owlia P, Fooladi AAI, Salimian J (2018a) An integrative in silico approach to the structure of Omp33-36 in Acinetobacter baumannii. Comput Biol Chem 72:77–86. https://doi.org/10.1016/j.compbiolchem.2018.01.003
Jahangiri A, Rasooli I, Owlia P, Imani Fooladi AA, Salimian J (2018b) Highly conserved exposed immunogenic peptides of Omp34 against Acinetobacter baumannii: an innovative approach. J Microbiol Methods 144:79–85. https://doi.org/10.1016/j.mimet.2017.11.008
Kumar A, Yogisharadhya R, Ramakrishnan MA, Viswas K, Shivachandra SB (2013) Structural analysis and cross-protective efficacy of recombinant 87 kDa outer membrane protein (Omp87) of Pasteurella multocida serogroup B: 2. Microb Pathog 65:48–56. https://doi.org/10.1016/j.micpath.2013.09.007
Lin L, Tan B, Pantapalangkoor P, Ho T, Hujer AM, Taracila MA, Bonomo RA, Spellberg B (2013) Acinetobacter baumannii rOmpA vaccine dose alters immune polarization and immunodominant epitopes. Vaccine 31:313–318. https://doi.org/10.1016/j.vaccine.2012.11.008
Luo G, Lin L, Ibrahim AS, Baquir B, Pantapalangkoor P, Bonomo RA, Doi Y, Adams MD, Russo TA, Spellberg B (2012) Active and passive immunization protects against lethal, extreme drug resistant Acinetobacter baumannii infection. PLoS ONE 7:e29446. https://doi.org/10.1371/journal.pone.0029446
McConnell MJ, Actis L, Pachón J (2013) Acinetobacter baumannii: human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol Rev 37:130–155. https://doi.org/10.1111/j.1574-6976.2012.00344.x
McConnell MJ, Pachón J (2010) Active and passive immunization against Acinetobacter baumannii using an inactivated whole-cell vaccine. Vaccine 29:1–5. https://doi.org/10.1016/j.vaccine.2010.10.052
Mendez JA, Soares NC, Mateos JS, Gayoso C, Rumbo C, Aranda J, Tomas M, Bou G (2012) Extracellular proteome of a highly invasive multidrug-resistant clinical strain of Acinetobacter baumannii. J Proteome Res 11:5678–5694. https://doi.org/10.1021/pr300496c
Moriel DG, Beatson SA, Wurpel DJ, Lipman J, Nimmo GR, Paterson DL, Schembri MA (2013) Identification of novel vaccine candidates against multidrug-resistant Acinetobacter baumannii. PLoS ONE 8:e77631. https://doi.org/10.1371/journal.pone.0077631
Ni Z, Chen Y, Ong E, He Y (2017) Antibiotic resistance determinant-focused Acinetobacter baumannii vaccine designed using reverse vaccinology. Int J Mol Sci 18:458. https://doi.org/10.3390/ijms18020458
Pachón J, McConnell MJ (2014) Considerations for the development of a prophylactic vaccine for Acinetobacter baumannii. Vaccine 32:2534–2536. https://doi.org/10.1016/j.vaccine.2013.10.064
Perez F, Bonomo RA (2014) Vaccines for Acinetobacter baumannii: thinking “out of the box”. Vaccine 32:2537–2539. https://doi.org/10.1016/j.vaccine.2014.03.031
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Rahbar MR, Rasooli I, Gargari SLM, Sandstrom G, Amani J, Fattahian Y, Jahangiri A, Jalali M (2012) A potential in silico antibody–antigen based diagnostic test for precise identification of Acinetobacter baumannii. J Theor Biol 294:29–39. https://doi.org/10.1016/j.jtbi.2011.10.026
Sefid F, Rasooli I, Jahangiri A, Bazmara H (2015) Functional exposed amino acids of BauA as potential immunogen against Acinetobacter baumannii. Acta Biotheor. https://doi.org/10.1007/s10441-015-9251-2
Singh R, Capalash N, Sharma P (2017) Immunoprotective potential of BamA, the outer membrane protein assembly factor, against MDR Acinetobacter baumannii. Sci Rep 7:12411. https://doi.org/10.1038/s41598-017-12789-3
Singh R, Garg N, Shukla G, Capalash N, Sharma P (2016) Immunoprotective efficacy of Acinetobacter baumannii outer membrane protein, FilF, predicted in silico as a potential vaccine candidate. Front Microbiol 7:158. https://doi.org/10.3389/fmicb.2016.00158
Solanki V, Tiwari V (2018) Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep 8:9044. https://doi.org/10.1038/s41598-018-26689-7
Song X, Zhang H, Zhang D, Xie W, Zhao G (2018) Bioinformatics analysis and epitope screening of a potential vaccine antigen TolB from Acinetobacter baumannii outer membrane protein. Infect Genet Evol 62:73–79. https://doi.org/10.1016/j.meegid.2018.04.019
Tsumoto K, Umetsu M, Kumagai I, Ejima D, Philo JS, Arakawa T (2004) Role of arginine in protein refolding, solubilization, and purification. Biotechnol Prog 20:1301–1308. https://doi.org/10.1021/bp0498793
Acknowledgements
The authors wish to thank Shahed University for supporting the conduct of this research. The authors appreciate the National Institute for Medical Research Development (NIMAD) Grant Number 965400.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research Involving Animals
All the experiments using animals in this work were conducted in Compliance with Ethical Standards and with animal care guidelines confirmed by Animal Care and Ethical Committee of Shahed University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rasooli, I., Abdolhamidi, R., Jahangiri, A. et al. Outer Membrane Protein, Oma87 Prevents Acinetobacter baumannii Infection. Int J Pept Res Ther 26, 2653–2660 (2020). https://doi.org/10.1007/s10989-020-10056-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-020-10056-0