Abstract
In many bacteria, the High Temperature requirement A (HtrA) protein functions as a chaperone and protease. HtrA is an important factor in stress tolerance and plays a significant role in the virulence of several pathogenic bacteria. Camostat, gabexate and nafamostat mesylates are serine protease inhibitors and have recently shown a great impact in the inhibition studies of SARS-CoV2. In this study, the inhibition of Listeria monocytogenes HtrA (LmHtrA) protease activity was analysed using these three inhibitors. The cleavage assay, using human fibrinogen and casein as substrates, revealed that the three inhibitors effectively inhibit the protease activity of LmHtrA. The agar plate assay and spectrophotometric analysis concluded that the inhibition of nafamostat (IC50 value of 6.6 ± 0.4 µM) is more effective compared to the other two inhibitors. Previous studies revealed that at the active site of the protease, these inhibitors are hydrolysed and one of the hydrolysates is covalently bound to the active site serine. To understand the mode of binding of these inhibitors at the active site of LmHtrA, docking of the inhibitors followed by molecular dynamics simulations were carried out. Analysis of the LmHtrA-inhibitor complex structures revealed that the covalently bound inhibitor is unable to occupy the S1 pocket of the LmHtrA which is in contrast to the previously determined camostat and nafamostat complex structures. This observation provides the first glimpse of the substrate specificity of LmHtrA which is not known. The obtained results also suggest that the development of novel inhibitors of LmHtrA and its homologs with active site architecture similar to LmHtrA can be pursued with suitable modification of these inhibitors. To date, only a very few studies have been carried out on identifying the inhibitors of HtrA proteolytic activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
High-Temperature Requirement A (HtrA), is a serine protease present in several Gram-negative bacteria in the periplasm. In Gram-positive bacteria, it is characterized as a surface-exposed protein. It is a critical stress response protein in many bacteria and is involved in ATP-independent protein quality control [1, 2].
Listeria monocytogenes has been reported as a human pathogen that induces listeriosis and several infections like meningitis and septicemia. HtrA of L. monocytogenes was identified as an important factor in stress tolerance and plays a significant role in the virulence potential of the bacterium [3]. It also promotes virulence in many other bacteria, including Campylobacter jejuni [4], Helicobacter pylori [5], Staphylococcus aureus [6], Streptococcus pneumonia [7] and Mycobacterium tuberculosis [8] by localizing to intracellular space and through direct secretion into the extracellular environment [9]. Hence, the search for inhibitors of HtrA is important for the control of infections.
The general architecture of HtrA consists of a central trypsin/chymotrypsin like protease domain followed by a PDZ (PSD-95, DLG1, and ZO-1) domain. A transmembrane domain in the N-terminal region is found only in Gram-positive bacterial HtrA’s whereas Gram-negative bacterial HtrA’s has a signal peptide and more than one PDZ domain. In our previous study with L. monocytogenes HtrA (LmHtrA), it was observed that LmHtrA interacts with extracellular matrix molecules and cleaves fibrinogen, plasminogen, fibronectin, and casein in a temperature and time-dependent manner [10]. In that study, it was found that the PDZ domain plays a significant role in substrate cleavage as native LmHtrA cleaves all tested substrates whereas LmHtrAΔPDZ cleaves only plasminogen and casein under certain conditions.
Camostat mesylate, Gabexate mesylate and Nafamostat mesylate are serine protease inhibitors and recently these compounds have shown a great impact in the inhibition studies of SARS-CoV2. Camostat is a potent trypsin protease inhibitor with an IC50 value of 50 nM [11]. It also inhibits other serine proteases such as TMPRSS2 [11], uPA [12], thrombin and hepsin [13], human prostasin, matriptase, airway trypsin-like protease (HAT) and β-tryptases [14]. It has been clinically used to treat chronic pancreatitis and has been reported as a potential therapeutic for SARS and MERS [15]. Camostat has also been studied in the inhibition of influenza viral replication and cytokine production [16]. It has been used in cell signaling studies and has antifibrotic, anti-inflammatory and potential antiviral activities [17]. The mesylate salt form of nafamostat is a broad-spectrum synthetic serine protease inhibitor, with anticoagulant, antiviral and antitumor activities [18, 19]. It is an effective inhibitor of human tryptase [20] and it blocks SARS-CoV2 infection of human lung cells with higher efficiency than camostat mesylate [21]. It has potential anti-viral and anti-inflammatory activities against COVID-19 and was reported as a safe and effective drug for COVID-19 and pneumonia [22]. Additionally, it is a good alternative for anticoagulation in ECMO [23]. A synthetic serine protease inhibitor, gabexate mesylate inhibits the invasion and metastasis of human colon cancer cells by blocking matrix metalloproteinases and neoangiogenesis [24]. This inhibitor is known for its anticoagulant property [25]. In a murine model of asthma, gabexate and nafamostat attenuate allergen induced airway inflammation and eosinophilia. Nafamostat and gabexate have been used to prevent pancreatitis related endoscopic retrograde cholangiopancreatography (ERCP) and in in vitro conditions nafamostat inhibits pancreatic protease activities 10–100 times more potently than gabexate mesylate [26].
In this study, the effect of these three inhibitors (camostat, gabexate and nafamostat mesylates) on the rLmHtrA protease activity against the substrates fibrinogen and casein was studied. Using azo-casein as a substrate, the half maximal inhibitory concentration (IC50) of these three inhibitors was calculated by a spectrophotometric method. Molecular docking and dynamics were performed to identify the mode of binding of the inhibitor with LmHtrA and the interacting atoms between the inhibitor and protein.
2 Materials and Methods
2.1 Expression and Purification of the rLmHtrA Protein
The L. monocytogenes gene encoding HtrA protein was cloned into the pET22b vector between BamHI and NdeI sites with a C-terminal His-tag which will aid in protein purification using Ni-NTA chromatography. The expression and purification of rLmHtrA were carried out as reported previously [10]. Briefly, the recombinant plasmid was transformed into the Escherichia coli expression host BL21(DE3) cells and for the protein expression, a single colony from the transformed plate was inoculated in 5 ml of Luria Bertani (LB) medium containing 100 µg/ml of ampicillin. The overnight grown bacterial culture at 37 °C, 150 rpm was then transferred into a mass culture of LB media (SRL, India) along with a 100 µg/mL concentration of ampicillin. After 2 h, the culture was induced with 1 mM IPTG and incubated at 37 °C with shaking of 150 rpm for about 3 h when an OD600 nm of 0.6 was obtained. The culture was centrifuged at 4000 rpm for 20 min and the pellet was lysed by sonication in a buffer containing 20 mM Tris pH 7.5, 200 mM NaCl, and 5% glycerol. The supernatant of the lysate was applied to a Ni-NTA affinity column and the bound protein was eluted using different concentrations of imidazole. The elution fractions were confirmed by SDS-PAGE gel analysis. The final purification was performed by size-exclusion chromatography and the prominent single peak of rLmHtrA protein obtained from the superdex-75 gel filtration column consisting of about six fractions of 1.0 ml were pooled, concentrated and used for the inhibition studies.
2.2 Determination of Protease Inhibitory Activity Using SDS-PAGE
Camostat mesylate, Gabexate mesylate and Nafamostat mesylate were obtained from Sigma Aldrich and TCI chemicals, India. Firstly, the native rLmHtrA (1 mg/mL) was incubated with different concentrations of inhibitors (1 to 25 mM) at 4 °C for 4 h, followed by the addition of substrate casein (1 mg/mL) or Fg (1.5 mg/mL) and the samples were incubated overnight at 37 °C. The rLmHtrA directly incubated with substrates and the rLmHtrA incubated with PMSF, the known serine protease inhibitor, and then incubated with substrates were used as controls. Finally, at the end of the incubation, the samples were analyzed using a 10% SDS-PAGE gel run at a constant voltage of 150 V and stained with Coomassie Brilliant Blue R-250. In the initial screening, nafamostat exhibited inhibition at 2 mM and hence further screening was carried out in the range of 0.15 mM to 2 mM.
2.3 Casein agar Plate Assay
The casein (Product No: 034021) was obtained from SRL, India. The casein agar plate assay was performed to confirm the inhibition of serine protease activity of LmHtrA. Casein was dissolved using NaOH and the pH was adjusted to 7.5. Agar 1% was added to the solution, which was then solidified in Petri plates.
In common, the central well contained trypsin (1 mg/ml). well 2 and 3 contained native rLmHtrA (1 mg/ml) and rLmHtrA with PMSF (positive control, 1 mg/mL) respectively. The surrounding wells 4, 5, and 6 contained rLmHtrA with different concentrations of inhibitors such as 5, 10, and 20 mM of camostat mesylate and 2, 5, and 10 mM of gabexate mesylate and 0.5, 1, and 2 mM of nafamostat mesylate incubated with rLmHtrA respectively. The casein agar plates were incubated at 37 °C for 18 h and the zone of inhibition was analyzed for each well.
2.4 IC50 Determination by UV-Vis Spectroscopy
For determining the IC50 value of the inhibitors, the inhibition assay was performed using the chromogenic substrate azo-casein (SRL, India). The azo-casein agar plate was first prepared, and the activity of the rLmHtrA against azo-casein was tested. Subsequently, the enzymatic action of rLmHtrA on azo-casein was measured spectrophotometrically and this was used as a control. For this experiment, azo-casein 5 mg/ml stock solution was prepared immediately before the assay, by dissolving azo-casein in sterilized water. 1 mg/ml of this solution was used for the assay along with 1.5 mg/ml purified rLmHtrA enzyme in 50 mM Tris-Buffer pH 8.0 containing 200 mM NaCl and 10 mM CaCl2. The protein-substrate mixture was incubated at 37 °C for 8 h and the reaction was terminated by the addition of 10% pre-cooled trichloroacetic, (TCA) solution. The solution was centrifuged at 10,000 rpm for 10 min and the 100 µl supernatant was transferred into a microcentrifuge tube followed by the addition of 0.5 M NaOH solution. The absorbance of the sample was measured using a UV-Visible spectrophotometer and a prominent peak was observed at 330 nm which indicates the presence of TCA-soluble azopeptides as a result of the enzymatic action of rLmHtrA on azo-casein.
To measure the IC50 value of the inhibitors, rLmHtrA (1.5 mg/mL) was preincubated with different concentrations of the inhibitors [camostat mesylate (1 nM to 20 mM), gabexate mesylate (1 nM to 10 mM), and nafamostat mesylate (1 nM to 5 mM)] at 4 °C for four hours before the addition of the substrate. To this enzyme-inhibitor mixture, 1 mg/ml of azo-casein was added and incubated at 37 °C for 8 h. As mentioned above, the reaction was terminated by the addition of TCA, and the solution was centrifuged. The absorbance at 330 nm was measured spectrophotometrically for the supernatant against appropriate blanks. The inhibition assay was repeated three times. The dose-response curves of enzyme inhibition by the three compounds were plotted and the IC50 value was calculated by fitting a nonlinear regression model to the data using GraphPad Prism 8 software.
2.5 Homology Modeling and Molecular Docking
The crystal structure of LmHtrA has not been determined. Therefore, homology modelling was performed using the SWISS-MODEL server. The LmHtrA sequence corresponding to the protease domain and PDZ domain was used for modeling. Using the crystal structure of DegQ from E. coli (PDB 3STJ) as a template, the LmHtrA structural model was obtained. The structure was validated using the SAVESv6.0 server (https://saves.mbi.ucla.edu/). In the modeled structure, some amino acids were found in the disallowed region of the Ramachandran map and these residues were manually modeled into the allowed region of the map.
For protein-ligand docking, initially the open-source software PyRx 0.8 [27] was used. First, to validate the docking protocol, the crystal structure of uPA (urokinase-type plasminogen activator) in complex with GBS (4-carbamimidamidobenzoic acid, cleaved camostat) (PDB 7DZD) was taken and the GBS molecule was manually removed. With apo-uPA structure the GBS was docked. The docking results were analyzed based on the binding affinity values and the ligand conformations were examined in comparison with the crystal structure of the uPA-GBS complex. The whole camostat molecule was also docked with uPA and compared with the crystal structure. Finally, to understand the mode of binding of camostat and gabexate with LmHtrA, the procedure used for uPA-GBS docking was repeated for the LmHtrA modelled structure.
2.6 Covalent Docking and Molecular Dynamics Simulation
The molecular docking of the cleaved part of camostat (4-carbamimidamidobenzoic acid, GBS) and gabexate (1-(5-carboxypentyl)-2 guanidinium) with the LmHtrA active site was performed using the CovDock program of Schrodinger. Maestro Protein preparation wizard was used for preparing the LmHtrA protein. Along with the default parameters, PROPKA at pH 7.0 was given for hydrogen bond assignments, and for all heavy atoms, the convergence criteria of 0.3 Å RMSD was considered. The OPLS_2005 force field was used for the minimization. The centroid of the active site residues (His229, Asp259 and Ser343) was selected as a docking site and Ser343 was selected as a reactive residue. While performing covalent docking, nucleophilic addition to a double bond reaction type and pose prediction docking mode was considered. In the final docked structure, the C7 atom of GBS / the C1 atom of 1-(5-carboxypentyl)-2 guanidinium is covalently bonded to the side chain oxygen of Ser343 and the model was visually inspected using Pymol.
Subsequently, LmHtrA-GBS and LmHtrA-1-(5-carboxypentyl)-2 guanidinium complex structures were subjected to molecular dynamics simulation using the Desmond package to understand the mode of interaction and stability of the docked GBS/1-(5-carboxypentyl)-2 guanidinium with LmHtrA. An orthorhombic box with a distance of 10 Å × 10 Å × 10 Å on the x, y and z-axis and the TIP3P water model was selected for solvation. The salt concentration of 0.15 M was given and the system was neutralized with Na + ions. A 50 ns molecular dynamics run was carried out in the NPT ensemble (temperature of 300 K, pressure of 1.01325 atm) with OPLS_2005 force field and the results were analyzed.
3 Results
3.1 Purification of rLmHtrA
The initial purification of rLmHtrA was carried out by Ni-NTA affinity chromatography. Following this, the purification was performed using size-exclusion chromatography. The peak fractions corresponding to rLmHtrA were pooled and concentrated and analyzed by 10% SDS-PAGE gel which is shown in Fig. 1. This purified protein was used for further studies.
3.2 Detection of Protease Inhibition Activity Using SDS-PAGE
The inhibition assay was performed to determine the effect of camostat, gabexate and nafamostat mesylates (Fig. 2) on the rLmHtrA protease activity. In order to find the minimum inhibitor concentration required for enzyme inhibition, the purified rLmHtrA was incubated initially with an increasing concentration of these inhibitors at 4 °C for 4 h and then the substrate was added to this enzyme-inhibitor solution. These samples were incubated overnight at 37 °C and finally analyzed using an SDS-PAGE gel. In our previous experimental studies, it was established that rLmHtrA was able to cleave the α chain of fibrinogen, β chain of casein, plasminogen and fibronectin [10]. Hence, casein (1 mg/ml) and human fibrinogen (Fg), (1.5 mg/ml) were selected as substrates for the present study.
The rLmHtrA (1 mg/ml) was incubated with Camostat mesylate (CM) from 2 mM to 25 mM increasing concentration. The substrate was then added to the enzyme-inhibitor mixture and incubated at 37 °C. The inhibition of rLmHtrA protease activity by CM against casein and fibrinogen is shown in Fig. 3A and B respectively. Without the inhibitor, rLmHtrA cleaves casein very efficiently (Lane 10). In the presence of the inhibitor, SDS-PAGE analysis revealed that at the low concentration of the inhibitor, partial inhibition was observed (3A, lane 4, 5 and 6) and at a concentration of 20 mM and 25 mM (lanes 7 and 8) a complete inhibition was observed.
Detection of protease inhibition activity using SDS-PAGE. Inhibition of rLmHtrA protease activity was demonstrated on a 10% SDS-PAGE gel using casein (A, C and E) and Fg (B, D and F) as substrates. Protein bands were visualized by Coomassie Brilliant Blue staining. In all gels, Lane 9 (rLmHtrA incubated with PMSF along with the substrate) and 10 (rLmHtrA incubated with substrates alone without the inhibitor) are control samples. Camostat mesylate (CM) was incubated with rLmHtrA and this complex was incubated with casein (A) and Fg (B). The cleavage of β-chain of casein and α chain of Fg are shown by a red arrow (Lane 10). With 20 mM of CM (lane 7), complete inhibition was observed as β-chain of casein and α chain of Fg found intact. Gabexate mesylate (GM) inhibits rLmHtrA against the substrates casein (C) and Fg (D). GM showed complete inhibition at a concentration of 5 mM (Lane 5), as no cleavage of β-chain of casein and α chain of Fg. Inhibition of rLmHtrA with nafamostat mesylate (NM) against the substrates casein (E) and Fg (F) respectively. NM was found to inhibit the cleavage of both substrates at a concentration of 1 mM (Lane 7). In A, C and E, the black arrow indicates the cleavage product of casein β-chain.
With regard to Fg (Fig. 3B), complete cleavage of the α chain of Fg was observed at the low inhibitor concentrations such as 2 mM, 5 mM and 10 mM (Lane 4, 5 and 6 respectively), whereas with 20 mM and 25 mM of CM, complete inhibition was observed as the three bands of Fg were clearly seen (Lane 7 and 8 respectively).
Figure 3C and D show the SDS-PAGE analysis of rLmHtrA activity in the presence of Gabexate mesylate (GM) with casein and Fg as substrates respectively. With 2 mM of GM incubated with rLmHtrA, the cleavage of casein (Fig, 3C, lane 4) and the α chain of Fg (Fig. 3D, lane 4) was observed whereas at 5 mM complete inhibition was observed (lane 5, Fig. 3C and D).
The initial experiments with Nafamostat mesylate (NM) concentrations in the range of 2 mM to 20 mM revealed that even at 2 mM the enzyme inhibition was observed. Subsequently, the experiment was conducted with NM concentrations in the range of 0.15 mM to 2 mM and the SDS-PAGE analysis indicated that at concentrations such as 0.15 mM, 0.25 mM, and 0.5 mM the cleavage of casein was observed (Fig. 3E, lanes 4, 5, and 6 respectively), whereas with 1 mM the inhibition was found (Fig. 3E, lane 7). The same observation was found with Fg (Fig. 3F). Lanes 9 and 10 correspond to the controls in all experiments, where rLmHtrA incubated with PMSF along with substrate showed complete inhibition (Lane 9) and rLmHtrA incubated with substrates without any inhibitor showed expected cleavage of casein and Fg (Lane 10, indicated by red arrow).
3.3 Casein agar Plate Inhibition Assay
The casein agar plate assay was also used to study the inhibition of rLmHtrA by CM (Fig. 4A), GM (Fig. 4B) and NM (Fig. 4C). In all the assays, the controls, trypsin (well 1) and rLmHtrA (well 2) were found to be proteolytically active and rLmHtrA with PMSF (well 3) showed complete inhibition of the enzyme activity. Wells 4, 5, and 6 correspond to increasing concentrations of the respective inhibitors. With CM, a complete inhibition, at 20 mM (Well 6) and a partial inhibition at 10 mM concentration (well 5) were prominently observed as seen in the SDS-PAGE analysis (Fig. 3A). With GM, 2 mM concentration showed the hydrolysis of casein (Fig, 4B, well 4) but 5 mM completely inhibited the rLmHtrA activity (Fig, 4B, well 5). Similarly with NM complete inhibition was observed at 1 mM concentration (Fig. 4C, well 5) and partial inhibition was observed at 0.5 mM concentration (Fig. 4C, well 4). Figure 4D shows the inhibition of all three inhibitors at their minimum inhibition concentration.
Casein agar diffusion plate assay to detect the proteolytic activity of rLmHtrA against the inhibitors with casein as a substrate. In common, well 1, 2, and 3: trypsin, rLmHtrA, and rLmHtrA incubated with PMSF respectively. Well 4, 5 and 6 increasing concentrations of the inhibitors (A): 5, 10 and 20 mM camostat incubated with rLmHtrA, in (B): 2, 5 and 10 mM gabexate incubated with rLmHtrA, in (C): 0.5, 1 and 2 mM nafamostat incubated with rLmHtrA and in (D): Inhibition of rLmHtrA with all three inhibitors, 20 mM camostat (well 4), 5 mM gabexate (well 5) and 1 mM nafamostat (well 6).
3.4 Protease Inhibition and IC50 Determination by UV-Vis Spectroscopy
The spectrophotometric inhibition assay was performed to determine the effect of CM, GM, and NM on the rLmHtrA protease activity and to calculate the IC50 value of the inhibitors. The azo-casein was used as a substrate in this assay. The rLmHtrA protease inhibition assay was performed by UV-Vis spectroscopy with appropriate blanks and controls as mentioned in the materials and methods section. As shown in Fig. 5, the inhibition assay results showed that CM, GM, and NM prominently inhibited rLmHtrA protease activity. Among these inhibitors, NM has the lowest IC50 value of 6.6 ± 0.4 µM, followed by GM and CM which have IC50 values of 80.56 ± 8.5 µM and 152.5 ± 7.5 µM respectively.
The inhibition of rLmHtrA protease activity by (A) Camostat (B) Gabexate and (C) Nafamostat. The dose-response curve was plotted and the curve fitting was done using GraphPad prism. Camostat, gabexate and nafamostat inhibit the activity of rLmHtrA protease with an IC50 value of 152.5 ± 7.5 µM, 80.56 ± 8.5 µM and 6.6 ± 0.4 µM respectively.
3.5 Binding of Inhibitors with rLmHtrA and their Interaction
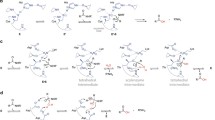
The classic catalytic triad of trypsin-like-serine protease contains His, Asp and Ser as amino acid residues. Based on the previous functional and crystallographic studies of serine proteases with camostat and nafamostat (PDB 3FVF [28]; 7DZD, [12]; 6ZOV [17]; 1RTK [29]; 7VM4, [30]; 7MEQ [31]), it is known that these molecules initially bind to the active site of the protease and get hydrolyzed at the central ester bond of the inhibitor. The cleaved product (4-guanidinobenzoic acid, termed GBS), which is common in camostat and nafamostat, covalently attaches to the side chain of an active site serine residue.
To understand the interaction between LmHtrA and camostat / gabexate, first the homology modelling of LmHtrA consisting of the serine protease domain and the PDZ domain was carried out (Fig. 6). Next, using the PyRx program, the protein-inhibitor docking was performed. Initially, to validate the docking protocol, the docking of GBS with the uPA structure was carried out. As found in the complex crystal structure of uPA-GBS, the ligand GBS was fitted into the S1 pocket of uPA (Fig. 7A). Subsequently, the docking of the whole camostat with uPA was carried out and this also showed that the positioning of the GBS moiety into the S1 pocket (Fig. 7B). Based on this, the docking of camostat and gabexate with LmHtrA was carried out. While analyzing the results, it is interesting to note that in the LmHtrA-camostat complex, the GBS moiety was unable to fit into the S1 pocket of the LmHtrA active site, which is in contrast with the other serine protease-GBS complexes mentioned above and also the docked structure of uPA-camostat. Examination of the S1 pocket of LmHtrA revealed that, the side chain of Lys361 projected into the S1 pocket and this possibly hindering the GBS from fitting inside. The sequence and structural comparison revealed that the corresponding position in other GBS complex structures (6ZOV, 3DZD and 3FVF) contains Gly residue (Figs. 6 and 8A). As a result, in other structures, the GBS molecule fits very tightly into the S1 pocket with its guanidinobenzoate moiety sandwiched between the two walls and the terminal NH2 group of the GBS interacts with an Asp residue at the bottom of the S1 pocket (Fig. 8B). Trypsin-like serine proteases usually cleave peptide bonds at Lys or Arg residues and this P1 residue fits snuggly into the S1 pocket. The bottom of the pocket contains a negatively charged conserved Asp residue which interacts with a positively charged P1 residue. This characteristic feature of a negatively charged residue at the S1 pocket is not present in LmHtrA and the Asp residue position is occupied by an alanine residue (Fig. 8A & C). With reference to the LmHtrA-gabaxate complex, like GBS, the 1-(5-carboxypentyl)-2 guanidinium group of gabexate is unable to fit into the S1 pocket. The mode of binding of camostat and gabexate at the LmHtrA active site is shown in Fig. 9A & B.
Sequence and structure comparison of LmHtrA with other GBS containing serine proteases. (A) The sequence of the LmHtrA serine protease domain is compared with the sequence of serine protease domain of Enteropeptidase (6ZOV), Urokinase-type plasminogen activator, uPA (7DZD) and Prostasin (3FVF). A very low sequence identity of 5.1%, 9.5% and 15.9% respectively is observed. However, the structures exhibit good conservation with an RMSD of 2.29 Å, 2.37 Å and 2.28 Å respectively for Cα atoms. Identical residues are shown in red. The blue arrow indicates the catalytic residues. The green box indicates the aspartic acid residue in the S1 pocket of 6ZOV, 7DZD and 3FVF whereas in LmHtrA it is alanine. The pink box indicates the Lys361 of LmHtrA which is projected into the S1 pocket whereas in other structures the corresponding position is glycine. (B) Structural superposition of 6ZOV, 7DZD and 3FVF showing the active site serine is covalently linked to GBS. (C). Active site region of LmHtrA. Lys361 is projected into the S1 pocket and the conserved Asp residue in the S1 pocket is replaced by A337 in LmHtrA.
Camostat and Gabexate at the active site of LmHtrA. Docking of Camostat (A) and Gabexate (B) at the active site of LmHtrA shows that the GBS of camostat and 1-(5-carboxypentyl)-2 guanidinium of gabexate are unable to fit into the S1 pocket. Mutation of Lys361 to glycine results in GBS (C) and 1-(5-carboxypentyl)-2 guanidinium (D) positioned inside the S1 pocket. Covalently linked GBS (E) and 1-(5-carboxypentyl)-2 guanidinium (F) interacting with β2 strand of LmHtrA. Both inhibitors are away from the S1 pocket due to the projection of Lys361 into the S1 pocket.
To validate the role of Lys361 in influencing the fitting of the ligand in the S1 pocket of LmHtrA, this residue was computationally mutated to Gly and the docking was performed with camostat and gabexate. As found in uPA and other serine proteases, in the mutated LmHtrA the GBS moiety of camostat and the 1-(5-carboxypentyl)-2 guanidinium group of gabexate fit into the S1 pocket (Fig. 9C & D).
To understand the interaction of the covalently linked GBS / 1-(5-carboxypentyl)-2 guanidinium group of gabexate with LmHtrA, covalent docking was performed for these two ligands at the active site of LmHtrA using the CovDock program of Schrödinger software. This resulted in a LmHtrA structure with Ser343 covalently linked to the ligands (GBS and 1-(5-carboxypentyl)-2 guanidinium). Followed by this, molecular dynamics simulation was performed for the LmHtrA complexes. In the LmHtrA-GBS complex, the guanidinobenzoate moiety is pointed opposite of the S1 pocket into the solvent environment (Fig. 9E) and at this position the primary amine nitrogen of the guanidino group interacts with the backbone carbonyl oxygen of Ser209 of a β2-strand that lines the active site cavity. Another interaction was found between the carbonyl oxygen of GBS and the backbone nitrogen of Gly341. In the case of the LmHtrA-1-(5-carboxypentyl)-2 guanidinium complex, the ligand that is covalently attached to Ser343 also points away from the S1 pocket into the solvent environment and is positioned similarly to GBS (Fig. 9F). As seen in GBS, its terminal primary amine nitrogens interact with the side chains of Asp233 and Asn190 and the imino nitrogen interacts with the side chain oxygen of Ser208 of β2-strand. Analysis of LmHtrA complex structures revealed that both inhibitors interact with the β2-strand of LmHtrA.
4 Discussion
The HtrA proteases of bacterial pathogens are attractive drug targets [32]. This objective is aimed at inhibiting their proteolytic activity. Towards this goal, initial studies in identifying anti-HtrA compounds were carried out with E. coli HtrA [33]. Following this, inhibition studies have been carried out with H. pylori HtrA (HpHtrA), Chlamydia trachomatis HtrA (CtHtrA) and a few small molecular compounds have been identified. For HpHtrA, through structure-based virtual screening, a small-molecule inhibitor termed HHI was identified which efficiently blocked E-cadherin cleavage with an IC50 value of 26 µM [5]. Using HHI as a template, further chemical modifications resulted in a new lead compound that exhibited a better IC50 value (13.2 µM) as compared to HHI [34]. Similarly for CtHtrA, two effective components were identified termed JO146 and JCP83. JO146 exhibited more potent antichlamydial activity than JCP83 [35]. Subsequently, by considering JO146 as a template, modifications were made at the P1, P2 and P3 positions and new analogues were generated; some of the compounds exhibited improved antichlamydial activity than the parent compound [36,37,38].
HtrA belongs to the serine protease family and the catalytic triad consists of Asp-His-Ser residues. Camostat, gabexate and nafamostat are serine protease inhibitors and they have been used as drugs clinically [11]. Recently, camostat and nafamostat have gained attention because of the studies related to the inhibition of TMPRSS2 of SARS-CoV-2 [31, 39]. In the present study, the inhibition of the proteolytic activity of LmHtrA was carried out with camostat, gabexate and nafamostat mesylates against the substrates fibrinogen and casein. The inhibition of LmHtrA was initially analyzed by SDS-PAGE gel which clearly showed that all the inhibitors effectively stopped the cleavage of casein and fibrinogen. Also, it was found that the concentration of nafamostat required for the inhibition is much lower than that of camostat and gabexate. Subsequently, casein agar plate inhibition assay was carried out and the results matched those of the SDS-PAGE analysis. To calculate the IC50 values of the inhibitors, a spectrophotometric inhibition assay was performed using azo-casein as a substrate. Nafamostat exhibited the lowest IC50 value of 6.6 ± 0.4 µM whereas for camostat and gabexate it is 152.5 ± 7.5 µM and 80.56 ± 8.5 µM respectively. Previous studies revealed that all three compounds undergo hydrolysis at the active site of the enzyme and part of the inhibitor covalently attaches to the serine residue at the active site. The crystal structures of serine proteases in complex with camostat and nafamostat revealed one of the hydrolysates of camostat/nafamostat, 4-guanidinobenzoic acid (GBS), covalently bound to the active site serine. Assuming LmHtrA exhibits a similar mechanism, to understand the interaction between the inhibitor and the LmHtrA active site residues, modelling of LmHtrA-inhibitor complexes was carried out and molecular dynamics simulations were done. The GBS of camostat/nafamostat and 1-(5-carboxypentyl)-2 guanidinium of gabexate, which are covalently linked to Ser343 of LmHtrA, unable to fit into the S1 pocket of LmHtrA which is in contrast with other GBS complex structures. As a result, in LmHtrA, the GBS and 1-(5-carboxypentyl)-2 guanidinium molecules adopt a different orientation and interact with the β2-strand of the core β-sheet. At this orientation, the inhibitors are more exposed to the solvent environment. As the inhibitors are away from the S1 pocket, this provides the opportunity for the addition of some suitable chemical groups at the terminal NH2 group of the inhibitors such that the modified component may make additional interactions with the protein which could provide more stability to the protein-inhibitor complex. Modifying the NH2 group of the GBS is not possible if the compound occupies the S1 pocket because of the space constraint. Therefore, for LmHtrA and its homologs with active site architecture similar to LmHtrA, the creation of novel inhibitors can also be investigated. In relation to this, in a previous study with CtHtrA, a peptidic inhibitor termed JO146 [Boc-Val-Pro-ValP(OPh)2] was initially identified as a potential inhibitor with an IC50 value of 12.5 µM and ~ 200 µM for peptide and protein based substrates respectively [35]. Later, to optimize the potency and selectivity of JO146, 23 analogues of the inhibitor were generated by changing residues at the P1 and P3 positions [36]. From this study, it was found that Val at the P1 position exhibited the best anti-chlamydial activity. Similarly, in HpHtrA inhibition studies using metal ions such as Zn++ and Cu++, it was found that Val, Ile and Ala were identified as the preferred amino acid residues in the P1 position [40]. The above two studies suggest a preference for smaller residues in the S1 pocket of the enzyme. These results coincide with our finding that in LmHtrA positioning of Lys361 at the bottom of the S1 pocket makes the pocket shallow and not suitable to accommodate bigger side chains. In CtHtrA and HpHtrA, the corresponding residue for Lys361 of LmHtrA is Ile265 and Ile239 respectively, and isoleucine could also make the S1 pocket shallow. In another study, considering JO146 as a lead compound, to improve the inhibitory activity of this molecule, 2-pyridone-based peptidomimetics were designed to replace the P2/P3 peptidic structure. Compared to JO146, these modified analogues exhibited improved selectivity over human neutrophil elastase by over 100-fold and inhibitory activity against CtHtrA by 5-fold [37]. In a recent study, P2 proline-modified analogues of JO146 were designed and one of the developed compounds exhibited potent anti-chlamydial activity in bacterial cell assays. The antiCtHtrA potency and selectivity of this compound over human neutrophil elastase improved by approximately 9- and 22-fold, respectively, as compared to JO146 [38].
LmHtrA is shown to cleave various host proteins [10]; however, the substrate specificity of the enzyme is not known. The position of Lys361 in LmHtrA and the inability of the GBS moiety to fit into the S1 pocket provide the first glimpse of the substrate specificity of LmHtrA. This provides the opportunity to design specific LmHtrA inhibitors as carried out for CtHtrA mentioned above. The crystal structure determination of rLmHtrA with camostat and gabexate will provide additional insights into the inhibition mechanism and inhibitor design.
References
Dong YK, Kyeong KK (2005) Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol 38:266–274. https://doi.org/10.5483/bmbrep.2005.38.3.266
Hansen G, Hilgenfeld R (2013) Architecture and regulation of HtrA-family proteins involved in protein quality control and stress response. Cell Mol Life Sci 70:761–775. https://doi.org/10.1007/s00018-012-1076-4
Abfalter CM, Bernegger S, Jarzab M et al (2019) The proteolytic activity of Listeria monocytogenes HtrA. BMC Microbiol 19:1–9. https://doi.org/10.1186/s12866-019-1633-1
Boehm M, Hoy B, Rohde M et al (2012) Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER: role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathog 4:1–12. https://doi.org/10.1186/1757-4749-4-3
Hoy B, Löwer M, Weydig C et al (2010) Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep 11:798–804. https://doi.org/10.1038/embor.2010.114
Rigoulay C, Entenza JM, Halpern D et al (2005) Comparative analysis of the roles of HtrA-like surface proteases in two virulent Staphylococcus aureus strains. Infect Immun 73:563–572. https://doi.org/10.1128/IAI.73.1.563-572.2005
Ibrahim YM, Kerr AR, McCluskey J, Mitchell TJ (2004) Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect Immun 72:3584–3591. https://doi.org/10.1128/IAI.72.6.3584-3591.2004
MohamedMohaideen NN, Palaninathan SK, Morin PM et al (2008) Structure and function of the virulence-associated high-temperature requirement A of Mycobacterium tuberculosis. Biochemistry 47:6092–6102. https://doi.org/10.1021/bi701929m
Xue RY, Liu C, Xiao QT et al (2021) HtrA family proteases of bacterial pathogens: pros and cons for their therapeutic use. Clin Microbiol Infect 27:559–564. https://doi.org/10.1016/j.cmi.2020.12.017
Radhakrishnan D, Amrutha MC, Hutterer E et al (2021) High temperature requirement A (HtrA) protease of Listeria monocytogenes and its interaction with extracellular matrix molecules. FEMS Microbiol Lett 368:1–11. https://doi.org/10.1093/femsle/fnab141
Shrimp JH, Kales SC, Sanderson PE et al (2020) An enzymatic TMPRSS2 assay for Assessment of clinical candidates and Discovery of inhibitors as potential treatment of COVID-19. ACS Pharmacol Transl Sci 3:997–1007. https://doi.org/10.1021/acsptsci.0c00106
Sun G, Sui Y, Zhou Y et al (2021) Structural basis of covalent inhibitory mechanism of TMPRSS2-Related serine proteases by Camostat. J Virol 95:e0086121. https://doi.org/10.1128/jvi.00861-21
Mahoney M, Damalanka VC, Tartell MA et al (2021) A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc Natl Acad Sci U S A 118:e2108728118. https://doi.org/10.1073/pnas.2108728118
Nimishakavi S, Raymond WW, Gruenert DC, Caughey GH (2015) Divergent inhibitor susceptibility among airway lumen-accessible tryptic proteases. PLoS ONE 10:1–17. https://doi.org/10.1371/journal.pone.0141169
Zhou Y, Vedantham P, Lu K et al (2015) Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 116:76–84. https://doi.org/10.1016/j.antiviral.2015.01.011
Yamaya M, Shimotai Y, Hatachi Y et al (2015) The serine protease inhibitor camostat inhibits influenza virus replication and cytokine production in primary cultures of human tracheal epithelial cells. Pulm Pharmacol Ther 33:66–74. https://doi.org/10.1016/j.pupt.2015.07.001
Sun W, Zhang X, Cummings MD et al (2020) Targeting enteropeptidase with reversible covalent inhibitors to achieve metabolic benefits. J Pharmacol Exp Ther 375:510–521. https://doi.org/10.1124/JPET.120.000219
Homma S, Hayashi K, Yoshida K et al (2018) Nafamostat mesilate, a serine protease inhibitor, suppresses interferon-gamma-induced up-regulation of programmed cell death ligand 1 in human cancer cells. Int Immunopharmacol 54:39–45. https://doi.org/10.1016/j.intimp.2017.10.016
Yan Y, Yang J, Xiao D et al (2022) Nafamostat mesylate as a broad-spectrum candidate for the treatment of flavivirus infections by targeting envelope proteins. Antiviral Res 202:105325. https://doi.org/10.1016/j.antiviral.2022.105325
Mori S, Itoh Y, Shinohata R et al (2003) Nafamostat mesilate is an extremely potent inhibitor of human tryptase. J Pharmacol Sci 92:420–423. https://doi.org/10.1254/jphs.92.420
Hoffmann M, Schroeder S, Kleine-Weber H et al (2020) Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob Agents Chemother 64:1–7. https://doi.org/10.1128/AAC.00754-20
Jang S, Rhee JY (2020) Three cases of treatment with nafamostat in elderly patients with COVID-19 pneumonia who need oxygen therapy. Int J Infect Dis 96:500–502. https://doi.org/10.1016/j.ijid.2020.05.072
Han SJ, Han W, Song HJ et al (2018) Validation of nafamostat mesilate as an anticoagulant in extracorporeal membrane oxygenation: a large-animal experiment. Korean J Thorac Cardiovasc Surg 51:114–121. https://doi.org/10.5090/kjtcs.2018.51.2.114
Yoon WH, Jung YJ, Kim TD et al (2004) Gabexate mesilate inhibits colon cancer growth, invasion, and metastasis by reducing matrix metalloproteinases and angiogenesis. Clin Cancer Res 10:4517–4526. https://doi.org/10.1158/1078-0432.CCR-04-0084
Yuksel M, Okajima K, Uchiba M, Okabe H (2003) Gabexate mesilate, a synthetic protease inhibitor, inhibits lipopolysaccharide-induced tumor necrosis factor-α production by inhibiting activation of both nuclear factor-κB and activator protein-1 in human monocytes. J Pharmacol Exp Ther 305:298–305. https://doi.org/10.1124/jpet.102.041988
Jae HC, In SL, Hyung KK et al (2009) Nafamostat for prophylaxis against post-endoscopic retrograde cholangiopancreatography pancreatitis compared with gabexate. Gut Liver 3:205–210. https://doi.org/10.5009/gnl.2009.3.3.205
Dallakyan S, Olson A (2015) Small- molecule Library Screening by docking with PyRx. Methods Mol Biol 1263:243–250. https://doi.org/10.1007/978-1-4939-2269-7
Spraggon G, Hornsby M, Shipway A et al (2009) Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations. Protein Sci 18:1081–1094. https://doi.org/10.1002/pro.118
Ponnuraj K, Xu Y, MacOn K et al (2004) Structural analysis of engineered bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell 14:17–28. https://doi.org/10.1016/S1097-2765(04)00160-1
Zhou Y, Wu J, Xue G et al (2022) Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat. Biophys J 18:3940–3949. https://doi.org/10.1016/j.bpj.2022.08.034
Fraser BJ, Beldar S, Seitova A et al (2022) Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation. Nat Chem Biol 18:963–971. https://doi.org/10.1038/s41589-022-01059-7
Wessler S, Schneider G, Backert S (2017) Bacterial serine protease HtrA as a promising new target for antimicrobial therapy? Cell Commun Signal 15:1–5. https://doi.org/10.1186/s12964-017-0162-533
Hauske P, Meltzer M, Ottmann C et al (2009) Selectivity profiling of DegP substrates and inhibitors. Bioorg Med Chem 17:2920–2924. https://doi.org/10.1016/j.bmc.2009.01.073
Perna AM, Rodrigues T, Schmidt TP et al (2015) Fragment-based De Novo Design reveals a small-molecule inhibitor of Helicobacter Pylori HtrA. Angew Chem Int Ed Engl 54:10244–10248. https://doi.org/10.1002/anie.201504035
Gloeckl S, Ong VA, Patel P et al (2013) Identification of a serine protease inhibitor which causes inclusion vacuole reduction and is lethal to Chlamydia trachomatis. Mol Microbiol 89:676–689. https://doi.org/10.1111/mmi.12306
Agbowuro AA, Hwang J, Peel E et al (2019) Structure-activity analysis of peptidic Chlamydia HtrA inhibitors. Bioorg Med Chem 27:4185–4199. https://doi.org/10.1016/j.bmc.2019.07.049
Hwang J, Strange N, Phillips MJA et al (2021) Optimization of peptide-based inhibitors targeting the HtrA serine protease in Chlamydia: design, synthesis and biological evaluation of pyridone-based and N-Capping group-modified analogues. Eur J Med Chem 224:113692. https://doi.org/10.1016/j.ejmech.2021.113692
Hwang J, Strange N, Mazraani R et al (2022) Design, synthesis and biological evaluation of P2-modified proline analogues targeting the HtrA serine protease in Chlamydia. Eur J Med Chem 230:114064. https://doi.org/10.1016/j.ejmech.2021.114064
Zhu H, Du W, Song M et al (2021) Spontaneous binding of potential COVID-19 drugs (Camostat and Nafamostat) to human serine protease TMPRSS2. Comput Struct Biotechnol J 19:467–476. https://doi.org/10.1016/j.csbj.2020.12.035
Bernegger S, Brunner C, Vizovišek M et al (2020) A novel FRET peptide assay reveals efficient Helicobacter pylori HtrA inhibition through zinc and copper binding. Sci Rep 10:1–13. https://doi.org/10.1038/s41598-020-67578-2
Acknowledgements
KP gratefully acknowledges the Science and Engineering Research Board (SERB), Government of India for the financial support in the form of a grant (CRG/2019/003342). AMC thanks SERB, India for the fellowship. The work of SW was supported by the grant I_4360 from the Austrian Science Fund (FWF).
Author information
Authors and Affiliations
Contributions
KP designed the study, analysed the data and wrote the manuscript. AMC experimental studies and the initial manuscript writing. SW provided the plasmid, analysed the results and manuscript correction.
Corresponding author
Ethics declarations
Conflict of interest
None declared.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
C, A.M., Wessler, S. & Ponnuraj, K. Inhibition of Listeria Monocytogenes HtrA Protease with Camostat, Gabexate and Nafamostat Mesylates and the Binding Mode of the Inhibitors. Protein J 42, 343–354 (2023). https://doi.org/10.1007/s10930-023-10114-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-023-10114-8