Abstract
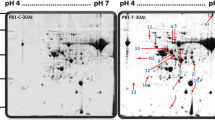
The resistant and susceptible genotypes of castor were utilized for leaf proteomic study during Fusarium wilt infection. The histopathological study was observed under SEM and it confirmed that the infection of Fusarium oxysporum f. sp. ricini was higher in the root of susceptible JI-35, while incompatible interaction is observed in resistant SKI-215 genotype. The acidic and neutral proteins were maximally up-expressed with 2 to 171 kDa in treated resistant and 2 to 150 kDa in treated susceptible interactions. In resistant genotype, the leaf proteins were recognized with 3.0- and 5.8-fold higher at infection stage and post-infection stage, respectively, as compared to susceptible genotype. The highly up expressions of leaf acidic (4.76 pI) and basic (8.77 pI) proteins were found with 224.94- and 61.68-fold change, respectively during the post-infection stage in treated resistance compared to its control. The protein spots at 4.76 pI and 8.77 pI were characterized with nanoLC–MS Triple TOF and were recognized as signalling molecules small GTP binding protein (23 kDa) and actin (8 kDa), respectively, on the basis of mass spectrometry and peptide sequences. However, basic and neutral proteins were up regulated as 30.11- and 20.30-fold changes in treated susceptible compared to its control. These proteins were identified as HSP90 (10 kDa) and LEA (27 kDa) proteins. The 148 kDa protein is recognized as histidine kinase in incompatible resistant interaction compared to compatible susceptible (serine threonine protein kinase, 65 kDa) as common acidic protein at 3.80 pI during infection stage. Some acidic proteins were maximally up-regulated in the leaf of resistant castor genotype and played a significant role in defense response.









Similar content being viewed by others
Data Availability
The data that support this study will be shared upon reasonable request to the corresponding author.
References
Ramanjaneyulu AV, Anudradha G, Ramana VM, Reddy AVV, Gopal NM (2017) Multifarious uses of castor (Ricinus communis L.). Int J Econ Plants 4:170–176
Salihu BZ, Gana AK, Gabadeyan T, Alabi MB (2014) Castor oil plant (Ricinus communis L.), a potential oil crop for agribusiness in Africa. Int J Appl Res Technol 3:29–35
Nanda S, Prasad N (1974) Wilt of castor a new record. Indian J Mycol Plant Pathol 4:103–105
Prasad MSL, Raoof MA, Gayatri B, Anjani K, Lavanya C, Prasad RD, Senthilvel S (2019) Wilt disease of castor, an overview. Indian Phytopathol 72:575–585
Mhaske SD, Mahatma MK, Jha S, Singh P, Ahmad T (2013) Polyamine metabolism and lipoxygenase activity during Fusarium oxysporum f. sp. ricini castor interaction. Physiol Mol Biol Plant 19:323–331
Andersen EJ, Ali S, Byamukam E, Yen Y, Nepal MP (2018) Disease resistance mechanism in plants. Genes 9:339–359
O’Farrell PH (1975) High resolution two dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021
Caruso G, Cavaliere C, Foglia P, Gubbiotti R, Samperi R, Lagana A (2009) Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Sci 177:570–576
Jangpromma N, Kitthaisong S, Lomthaisong K, Daduang S, Jaisil P, Thammasirirak S (2010) A proteomics analysis of drought stress responsive proteins as biomarker for drought tolerant sugarcane cultivars. Am J Biochem Biotechnol 6:89–102
Panera A, Jalu RK, Virani HB (2018) Inheritance of Fusarium wilt resistance and certain morphological characters in castor (Ricinus communis L.). Bull Environ Pharmacol Life Sci 7:30–35
Priya PB, Kumar MVN, Shankar VG (2016) Identification of potential sources of resistance for wilt caused by Fusarium oxysporum f. sp. ricini in castor (Ricinus communis L.). Asian J Biosci 11:284–288
Joshi NS, Rao KS, Subramanian RB (2012) Anatomical and biochemical aspects of interaction between roots of chickpea and Fusarium oxysporum f. sp. ciceris race 2. Arch Phytopathol Plant Prot 1:1–17
Damerval C, Devienne D, Zivy M, Thiellement H (1986) Technical improvement in two dimensional electrophoresis increase the level of genetic variation detected in wheat seedling proteins. Electrophoresis 7:52–54
Ashoub A, Berberich T, Beckhaus T, Bruggemann W (2011) A competent extraction method of plant proteins for 2D gel electrophoresis. Electrophoresis 32:2975–2978
Nandakumar MP, Marten MR (2002) Comparison of lysis methods and preparation protocols for one- and two-dimensional electrophoresis of Aspergillus oryzae intracellular proteins. Electrophoresis 23:2216–2222
Kausar R, Arshad M, Shahzad A, Komatsu S (2012) Proteomics analysis of sensitive and tolerant barley genotypes under drought stress. Amino Acids 44:345–359
Silva TD, Almeida CMA, Malafaia CB, Oliveira LMS, Silva MV, Correia MTS (2017) Analysis of protein profile of tomato root infected with Fusarium oxysporum f. sp. lycopersici. Genet Mol Res 16:1–10
Eggert K, Zorb C, Muhling KH, Pawelzik E (2011) Proteome analysis of Fusarium infection in emmer grains (Triticum dicoccum) Plant Pathol 60:918–928
Xue R, Wu J, Zhu Z, Wang L, Wang X, Wang S, Blair MW (2015) Differentially expressed genes in resistant and susceptible common bean (Phaseolus vulgaris L.) genotypes in response to Fusarium oxysporum f. sp. phaseoli. PLoS ONE 10:1–20
Campos FAP, Nogueira FCS, Cardoso KC, Costa GCL, Bem LEV, Domont GB, Da Silva MJ, Moreira RC, Soares AA, Juca TL (2010) Protein analysis of castor bean seeds. Pure Appl Chem 82:259–267
Wang L, Huang Y, Wang X, Chen Y (2019) Label-free LC–MS/MS proteomics analyses reveal proteomic changes accompanying MSTN KO in C2C12 cells. BioMed Res Int. https://doi.org/10.1155/2019/7052456
Antala TJ, Gajera HP, Mandavia MK, Dave RA, Kothari VV, Golakiya BA (2015) Leaf proteome alterations in tolerant pearl millet (Pennisetum glaucum L.) genotype under water stress. IJAEB 8:539–549
Yang H, Huang Y, Zhi H, Yu D (2011) Proteomics based analysis of novel genes involved in response toward soybean mosaic virus infection. Mol Biol Rep 38:511–521
Geddes J, Eudes F, Laroche A, Selinger BL (2008) Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host, Hordeum vulgare. Proteomics 8:545–554
Zhou W, Eudes F, Laroche A (2006) Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 6:4599–4609
Li X, Bai T, Li Y, Ruan X, Li H (2013) Proteomic analysis of Fusarium oxysporum f. sp. cubense tropical race 4-inoculated response to Fusarium wilts in the banana root cells. Proteome Sci 11:41–55
Ma QH (2007) Small GTP-binding proteins and their functions in plants. J Plant Growth Regul 26:369–388
Day B, Henty JL, Porter KJ, Staiger CJ (2011) The pathogen–actin connection, a platform for defense signaling in plants. Annu Rev Phytopathol 49:483–506
Leontovycova H, Kalachova T, Trda L, Pospichalova R, Lamparova L, Dobrev PI, Malinska K, Burketova L, Valentova O, Janda M (2019) Actin depolymerization is able to increase plant resistance against pathogens via activation of salicylic acid signalling pathway. Sci Rep 9:10397
Xu ZS, Li ZY, Chen Y, Chen M, Li LC, Ma YZ (2012) Heat shock protein 90 in plants, molecular mechanisms and roles in stress responses. Int J Mol Sci 13:15706-15723
Bo SH, Suo LZ, An SM (2005) LEA proteins in higher plants, structure, function, gene expression and regulation. Colloids Surf B 45:131-135
Nongpiur R, Soni P, Karan R, Pareek SL, Pareek A (2012) Histidine kinases in plants, cross talk between hormone and stress responses. Plant Signal Behav 10:1230-1237
Hardie DG (1999) Plant protein serine/threonine kinases, classification and functions. Annu Rev Plant Physiol Plant Mol Biol 50:97-131
Acknowledgements
Any specific fund did not receive for the research.
Funding
Any specific fund did not receive for the research.
Author information
Authors and Affiliations
Contributions
HJK carried out the experiment, microbial work, biochemical analysis, and worked out the results; HPG was responsible for interpretation of data, writing of the manuscript and designing the experiments. DRM was liable for the idea and coordination of the experiment. DGH and RVB responsible for helping in microbial and biochemical analyses; and analyzed the data. RAD carried out nanoLC–MS analysis. All authors contributed critically to the drafts and gave final approval for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kachhadiya, H.J., Gajera, H.P., Mehta, D.R. et al. Comparative Appraisal of Leaf Proteomic and Mass Spectrometry Analyses During Fusarium Wilt Infection in Resistance and Susceptible Genotypes of Castor (Ricinus communis L.). Protein J 41, 638–658 (2022). https://doi.org/10.1007/s10930-022-10083-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-022-10083-4