Abstract
Moloney murine leukemia virus reverse transcriptase (MMLV-RT) is the most frequently used enzyme in molecular biology for cDNA synthesis. To date, reverse transcription coupled with Polymerase Chain Reaction, known as RT-PCR, has been popular as an excellent approach for the detection of SARS-CoV-2 during the COVID-19 pandemic. In this study, we aimed to improve the enzymatic production and performance of MMLV-RT by optimizing both codon and culture conditions in E. coli expression system. By applying the optimized codon and culture conditions, the enzyme was successfully overexpressed and increased at high level based on the result of SDS-PAGE and Western blotting. The total amount of MMLV-RT has improved 85-fold from 0.002 g L−1 to 0.175 g L−1 of culture. One-step purification by nickel affinity chromatography has been performed to generate the purified enzyme for further analysis of qualitative and quantitative RT activity. Overall, our investigation provides useful strategies to enhance the recombinant enzyme of MMLV-RT in both production and performance. More importantly, the enzyme has shown promising activity to be used for RT-PCR assay.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Reverse transcriptase (RT) has been initially discovered in 1970 and isolated from retrovirus (tumor viruses) [1, 2]. The name of retrovirus originally derives from the capability of these viruses to perform the replication in host cells by converting their RNA genomes into DNA. The process which is called reverse transcription naturally occurred due to the presence of RT enzymes [3]. Briefly, RT is an RNA-dependent DNA polymerase that converts a sequence of single-stranded RNA as a template into a sequence of complementary DNA (cDNA) as a product [4]. The source of RT can be found in human immunodeficiency virus (HIV), Moloney murine leukemia virus (MMLV), avian myeloblastosis virus (AMV), and other retrovirus [5]. Generally, RT is a monomeric or dimeric protein that has two active sites consisting of both DNA polymerase and RNase H endonuclease [6].
The discovery of RT has revolutionized modern molecular biology and led to the revision of central dogma, in which the alteration of DNA into RNA becomes a reversible step. The finding has also encouraged scientists to evolve advanced research in transcriptomics [7]. In addition, the enzyme plays an important role as a molecular tool in RT-PCR, RNA sequencing, analysis of gene expression, cDNA cloning, and any other molecular approach that involves the synthesis of cDNA from RNA molecules [8]. Moreover, RT is responsible for the first step procedure in RT-PCR. In the 2019 coronavirus disease (COVID-19) pandemic era, the use of the enzyme in RT-PCR has been essential and crucial as the approach is considered to be the gold standard for the COVID-19 diagnostic test [9, 10]. The RT from MMLV is the most extensively used and preferred in molecular research or laboratory works due to its high catalytic activity and fidelity, thus it is commercially valuable and promising [11]. For those reasons, MMLV-RT has been further investigated in this study.
Previous studies have reported that the thermostability and efficiency of MMLV-RT can be improved by utilizing some strategies such as site-directed mutagenesis, rational design, and recombinant enzyme production using E. coli expression system [12, 13]. Potent RNase H activity is beneficial in PCR application to degrade RNA in RNA-DNA duplex during the first cycles of PCR. On the contrary, with long RNA templates, RNase H activity may early degrade RNA resulting in truncated cDNA. Therefore, low RNase H activity has the advantage to produce good quality of long transcripts in cDNA amplification [14]. By reducing RNase H activity, MMLV-RT has been regarded to be more thermostable. In consequence, non-specific binding of primers during amplification and RNA secondary structure can be minimized [15].
The present study attempts to develop the MMLV-RT using a synthetic gene by employing E. coli strain BL21 star as an expression host. The protein sequence of wild-type MMLV that encodes RT has been modified to decrease RNase H activity. The substitution of amino acids has been made and positioned at Y139A, T197E, and F139N according to the earlier study by Potter and Rosenthal [16]. The study has focused on combining the optimization of codon and culture conditions in order to seek effective ways of boosting recombinant MMLV-RT production in E. coli. The protein obtained has been purified and applied for RT-PCR assay to observe its performance and activity. Hence, this study was objected to supposedly find optimum conditions and produce the highest content and activity of purified RT in the laboratory scale.
2 Materials and Methods
2.1 Bacterial Strain, Plasmid, and Medium
The host strain for protein expression used in this study was Escherichia coli BL21 Star (DE3) (Invitrogen). Plasmid pD451 for expression vector was synthesized by ATUM, Inc (Newark, CA) harboring reverse transcriptase gene from MMLV and containing isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible T7 promoter, ori pUC, and kanamycin antibiotic marker. Luria-Bertani (LB) medium was purchased from Sigma-Aldrich (USA).
2.2 Design of Synthetic Gene Encoding MMLV-RT
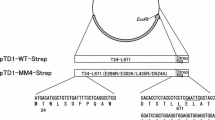
A gene sequence encoding MMLV-RT was designed according to US Patent No 8541219 B2 [16]. The target protein has three mutations (Y139A, T197E, and F139N) and consists of 504 aa. In the construction of the expression cassette, a 6 × Histidine tag was added at the N-terminal of MMLV-RT sequence followed by the enterokinase cleavage site (Fig. 1). The solubility of the target protein was determined using SOLUPROT v1.0 followed by disulfide bond analysis using DISULFIND software [17, 18]. The full-length construct of his-MMLV-RT sequence was then subjected to Gene Designer software for codon optimization according to E. coli codon usage (performed by ATUM, Inc). The resulting gene sequence has been analyzed using other software to get a more optimum codon sequence. Codon adaptation index (CAI) and % GC of the gene sequences was calculated using CAI/cal [19]. The mRNA folding energy profile near the translation initiation region (TIR) was observed using RNAfold and RNAstructure [20]. After that, the codon sequence was reoptimized using the IDT codon optimization tool to get the desired codon sequence [21]. Subsequently, the codon-optimized sequence was translated into protein using the Expasy translate tool and aligned with the initial template using Clustal Omega to check the mutation [22, 23]. Lastly, the codon-optimized gene encoding MMLV-RT was synthesized, sequenced-verified, and cloned into pD451-SR containing T7 promoter by ATUM, Inc (Newark, CA).
2.3 Transformation of MMLV-RT Plasmid into E. coli BL21
The constructed plasmid, pD451-SR_MMLV-RT, containing gene encoding M-MLV RT was transformed into E. coli BL21 star (DE3) using PEG method. The PEG method was performed following protocol described by Chung et al. with several modifications [24]. Transformants were plated on LB agar plates containing 30 mg L−1 kanamycin and the plates were incubated at 37 °C for 16 h. Transformant colonies on LB-kanamycin agar plates were selected for further colonies screening.
2.4 Expression and Optimization of Induction Conditions for Recombinant Protein Expression
To achieve the optimum conditions for the expression of MMLV-RT, variations of temperature, inducer concentration, post-incubation time, and pre-induction optical density were performed. The expression of the recombinant protein upon varying conditions was analyzed using SDS-PAGE.
2.4.1 Variation of Temperature
Upon transformation of the plasmid to the E. coli BL21 Star (DE3) strain, single bacterial colonies were selected from the transformants and inoculated into 5 mL of LB medium supplemented with 30 mg L−1 kanamycin (LB-kanamycin) and 0.4% glucose. The pre-culture was grown for 16 h at 37 °C, 165 rpm. A volume of 1% of overnight pre-culture was inoculated to 22 mL of LB-kanamycin at 37 °C until the culture reached the OD600 of 0.8–1. Subsequently, the culture was induced with 0.2 mM of IPTG. After the addition of the inducer, for the temperature optimization, cultures were incubated at 18 and 37 °C overnight.
2.4.2 Variation of Inducer Concentration
A volume of 1% overnight pre-culture was inoculated in 22 mL of LB-kanamycin medium. After reaching the OD600 of 0.8–1, cultures were induced with various concentrations of IPTG (0.1, 0.2, 0.4, 0.6, 0.8, and 1 mM) and subsequently incubated at the chosen temperature overnight.
2.4.3 Variation of Post-induction Incubation Time
A further step was to vary the post-induction incubation time. Once an OD600 reached at 0.8–1.0, cultures were induced with IPTG, which imposed the highest protein concentration and incubated at 18 °C. The time of incubation varied at 6, 12,18, 24, 30, and 36 h.
2.4.4 Variation of Pre-induction Optical Density
Final experiment was to optimize the optical density of the culture. Cultures were grown until the OD600 reached 0.4, 0.6, 0.8, and 1. The analysis of expression for each of OD600 variations was performed. All optimized parameters obtained from this step were applied for further investigation.
2.5 Analysis of Expression by SDS-PAGE
The enzyme expression was observed using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) [25]. The treatment of cell pellets at the end of cultivation for each of the expression conditions was performed according to Larentis et al. with modifications [26]. Briefly, at the end of cultivation, the culture was transferred to 50 mL conical flask and cells were harvested by centrifugation at 8000 rpm for 6 min at 4 °C. The cell pellet was prepared in 25 mM Tris-HCl buffer (pH 8) and placed on ice during cell disruption by sonication. Afterwards, the suspension was then centrifuged at 14,000 rpm for 15 min at 4 °C to obtain the cell-free extract (soluble fraction). For the variation of temperature, the total (fractions directly obtained after sonication, prior to centrifugation of suspension), soluble, and insoluble fractions were subjected to SDS-PAGE, while only the soluble fractions of the remaining variations were subjected to SDS-PAGE. Protein bands were stained with Coomassie Brilliant Blue and the concentration was determined using bovine serum albumin (BSA) as a standard. The protein band was then analyzed using ImageJ and the content of that in each variation was estimated by comparing the area under curved (AUC) of samples to the standard.
2.6 Expression and Purification of Recombinant MMLV-RT
E. coli BL21 Star (DE3) harboring pD451-SR_MMLV-RT was grown for pre-culture in LB-kanamycin medium containing 0.4% glucose at 37 °C with shaking at 165 rpm for 16 h. The 1% of pre-culture (220 µl) was inoculated to 22 mL LB-kanamycin medium, and the culture was incubated at 37 °C with shaking at 165 rpm. When the absorbance of culture reached 1.0 at 600 nm, 0.2 mM IPTG was added to induce the synthesis of MMLV-RT. The overexpression of MMLV-RT was performed overnight at 18 °C with shaking at 165 rpm. The cells were then harvested by centrifugation at 4 °C for 6 min at 8000 rpm and resuspended in 50 mM Tris-HCl buffer (pH 8.0). The resuspended cells were disrupted by sonication and the cell debris was cleared by centrifugation at 4 °C for 15 min at 14,000 rpm. The obtained supernatant was loaded onto a HisTrap™ HP (1 mL; Cytiva) column equilibrated with 20 mM sodium phosphate (pH 7.4) containing 500 mM NaCl and 20 mM imidazole. The bound protein was gradually eluted with 20 mM up to 500 mM imidazole in a 20 mM sodium phosphate buffer (pH 7.4) containing 500 mM NaCl. The eluted fractions were pooled together and dialyzed against 50 mM Tris-HCl buffer (pH 8.0) to remove imidazole. The first dialysis was performed at 4 °C for 6 h and continued with the second dialysis at the same temperature overnight. The purity of MMLV-RT was evaluated by SDS-PAGE on a 10% polyacrylamide gel and the protein concentration was examined in accordance with the previous method [27].
2.7 Western Blotting
Purified recombinant MMLV-RT protein was applied onto acrylamide gel, then transferred to nitrocellulose membranes using Mini Protean® II trans blot unit (Bio-Rad). The membrane was blocked with BSA/TBST solution for 1 h at room temperature with shaking. The membrane was washed two times with TBST solution for 10 min each and incubated with HisProbe-HRP (Thermo Scientific, US) working solution for 1 h with shaking. After that, the washing step was repeated four times, and detection was performed by adding KPL TMB Peroxidase substrate directly to the membrane.
2.8 Activity Assay
For qualitative assay, viral RNA of SARS-CoV-2 was used as a template. A two-step RT-PCR assay consisting of reverse transcription and PCR was performed according to the ARTIC nCoV-2019 sequencing protocol V3 (LoCost) [28]. Firstly, the complementary DNA (cDNA) was synthesized using Lunascript-No RT supermix (New England Biolabs/NEB) added with SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) as a positive control. Meanwhile, a negative control was prepared using Lunascript-No RT supermix only and samples were treated by adding Lunascript-No RT supermix with 2 µL of purified MMLV-RT, respectively. All reactions for cDNA synthesis were carried out in 10 µL reaction mixtures and then incubated as follows: 25 °C for 2 min, 55 °C for 10 min, 95 °C for 1 min, and hold at 4 °C. The cDNA obtained was amplified using Q5 Hot Start High-Fidelity Master Mix (NEB) and V3 primers (IDT) to generate overlapping 400 bp (bp) amplicons covering the SARS-CoV-2 genome. The PCR reaction was set up for 30 cycles. All PCR products were run in 1.2% agarose gel using an electrophoresis instrument and visualized using a UV Transilluminator.
For quantitative assay, The RT activity was measured using EnzChek RT assay kit (Invitrogen) following the manufacturer’ instruction. All standards and samples were applied in 5 µL of each reaction. Serial dilutions of commercial MMLV-RT (SSIV, Thermo Fisher Scientific) were used as standards. The reaction was stopped with 200 mM EDTA and DNA-RNA duplex obtained from a mixture of poly(A) template, oligo-dT primer and dTTP was detected by the PicoGreen dye. The RT activity was determined by fluorescence intensity using a microplate reader with standard wavelengths of excitation and emission at 480 and 520 nm, respectively.
3 Results
3.1 Design of Bene Encoding MMLV-RT
According to US Patent No 8541219 B2, the mutant variant of MMLV-RT that has been chosen in this study has reduced RNase H activity compared to the native. The solubility of the target protein was determined using SOLUPROT and gave a score of 0.873 of 1, indicating that the protein was compatible with expression in the E. coli system. This result was strengthened by DISULFIND software analysis which showed no intramolecular disulfide bond formation inside the target protein (data not shown). The amino acid sequence of the target protein was then used as a template for designing the synthetic gene. Even though both DNA and amino acid sequence can be used as a template, the latter was preferred because the target protein is an unnaturally found mutant variant.
Improving heterologous protein expression through codon optimization can increase the commercial value of its recombinant protein product. Codon optimization was done by replacing the native codon with E. coli codon usage. Only synonymous codon usage within the open reading frame was varied. The initial codon-optimized sequence was generated by Gene Designer. Even though the CAI value is good, the result of the gene’s parameter analysis using CAI/cal showed that the %GC of the third nucleotide in the codon is still high (Table 1). Because it is fundamental in mRNA secondary structure formation, the value must be lowered until it is close to the %GC3 of E. coli listed on the codon usage table (Kazusa). Sequence re-optimization was carried out by randomly picking the AT-rich codon except for the rare codon using IDT codon optimization tool. As a result, the %GC3 was decreased from 66.7 to 59.7% which is closer to the requirement of E. coli %GC3 (57.23%).
The mRNA folding energy of near TIR is also controlled using RNAfold and RNAstructure software. Table 2 showed the nucleotide sequence used for mRNA folding analysis. The result from RNAfold and RNAstructure showed that the initial mRNA folding energy values were low enough to produce spontaneous mRNA folding formation (Table 1). To increase the mRNA folding energy, the synonymous codon substitution was performed. The result showed the increase of mRNA folding energy which means the unstable mRNA secondary structure (Table 1).
The final sequence should be confirmed to ensure there was no mutation at the protein level after the codon optimization process. ExPASy translate tool was used to convert DNA sequence into amino acids (Supplementary Fig S1). Then, the translated protein was aligned with the initial template using Clustal omega and the result showed no mutation has occurred (Supplementary Fig S2).
3.2 Expression and Induction Conditions for Optimal Recombinant Protein Expression
3.2.1 Post-induction Temperature
Prior to varying the culture conditions, the expression of MMLV-RT was performed under the condition of 37 °C, using 0.2 mM IPTG at the OD of 0.8–1, and incubated overnight. Expression and solubility of MMLV-RT was analyzed using SDS-PAGE as depicted in Fig. 2. At 37 °C, the recombinant enzymes were overexpressed in its insoluble fraction. This result indicated that the host cell formed inclusion bodies. To obtain a more soluble recombinant enzyme, we varied culture conditions, including post-induction temperature at 18 °C, 27 °C, and 37 °C. The estimated protein yield for each variation is displayed in Fig. 3. As indicated, the culture shows a higher level of protein yield suggesting significant improvement of its solubility at 18 °C. The protein yield for soluble fraction (0.065 g L−1) was 32 times higher than that at 37 °C (0.002 g L−1), and twice higher compared to protein at 27 °C. This result is in agreement with the SDS-PAGE of total, soluble, and insoluble fractions, depicted in Fig. 2. At 18 °C, it was observed that the targeted protein was expressed more in soluble fraction than that at 37 °C and 27 °C.
In terms of bacterial biomass, the optical density at the end of cultivation for all tested temperatures were quite similar (5.63, 5.85, and 5.51 at 37 °C, 27 °C, and 18 °C, respectively). Based on the above results, we selected a post-incubation temperature of 18 °C as the preferred temperature for further studies.
3.2.2 IPTG Concentrations
The concentration of IPTG was varied to seek the optimum IPTG concentration to induce MMLV-RT. The concentration ranges used were from 0.1 to 0.8 mM of IPTG and no inducer was applied for the negative control. The estimated protein yield for each of the variations was shown in Fig. 4. According to Fig. 4, IPTG concentration of 0.1 mM is sufficient to boost the MMLV-RT expression (0.074 g L− 1). Apparently, in the range of 0.1–0.8 mM, the protein yield obtained was not significantly different. Increasing the IPTG concentration to more than 0.1 mM did not improve the yield.
As for the biomass, based on Fig. 4, it can be observed that the optical densities of induced cultures showed no significant differences. Considering the results, 0.1 mM was the most suitable concentration of IPTG, thus it was selected for promoting MMLV-RT expression.
3.2.3 Post-induction Incubation Time
The effect of diverse post-induction incubation time was also studied. We varied the incubation time from 6 to 24 h. The estimated protein yield increased as the time of incubation was prolonged (Fig. 5). The yield upon 24 h of post-incubation time reached the highest with 0.083 g L−1. This value was 1.3-3 times higher than those obtained in other incubation times. Extending the incubation to over 24 h was not required as this led to the lowering of protein yield (data not shown).
Optical densities of the cultures were also observed for each post-induction time. According to Fig. 5, the values follow the same trend as the protein yield. The biomass increased as the incubation time got prolonged and reached maximum at 24 h. Considering the results, 24 h of incubation was selected as the optimum post-induction incubation time.
3.2.4 Initial Pre-induction Optical Density (OD600)
The OD of culture at initial pre-induction was varied. Cultures were incubated until they reached the OD of 0.4–2. As it can be observed from Fig. 6, induction performed at the OD of 0.4 is sufficient to obtain the highest estimated protein yield of MMLV-RT. There was no significant increase upon inducing the culture at the OD of 0.8, while induction performed at the OD of 1 and 2 lowered the result.
In addition to protein yield, we also observed the optical densities of the cultures to measure bacterial biomass. The same trend was observed, as the induction performed at OD of 0.4–0.8 resulted in similar biomass at the end of cultivation. However, induction at the OD of 1–2 led to lower bacterial biomass. Considering both the yield and bacterial biomass, we chose to employ the induction at OD 0.4 as the preferred condition. The protein yield obtained when performing the induction in the current condition (0.175 g L−1) was apparently improved 85 times than that obtained upon employing the initial condition (0.002 g L−1).
3.3 Purification of Recombinant MMLV-RT
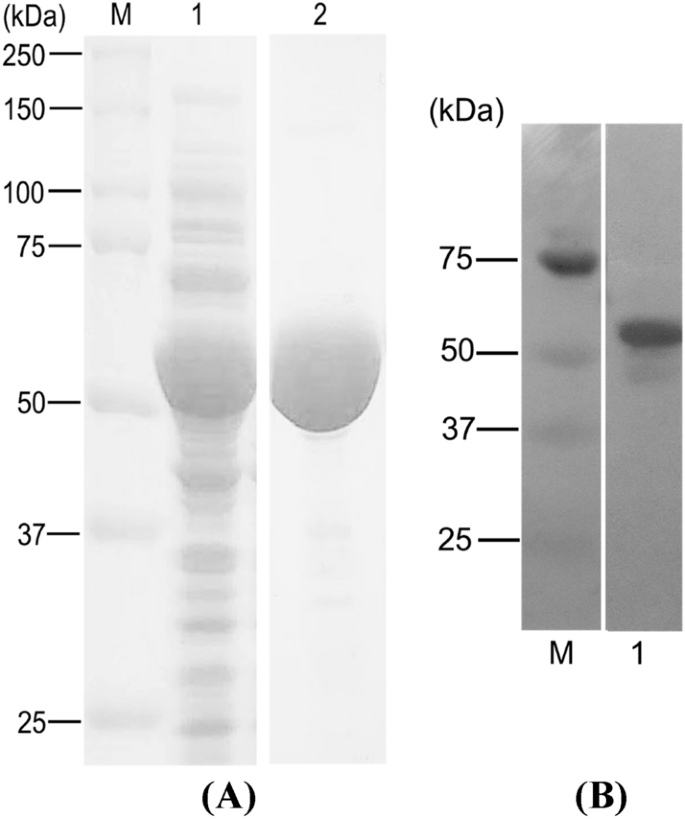
Purification of MMLV-RT was performed to obtain quite a high purity of the enzymes before enzyme activity assay. The enzyme was overexpressed in E. coli BL21 star (DE3) in LB-kanamycin. The purified enzyme was obtained after purification by Ni-affinity chromatography (Ni Sepharose® excel). The purity of the enzyme was analyzed using SDS-PAGE. A high purity MMLV-RT was successfully obtained with the molecular mass at 58 kDa, in accordance with the molecular weight predicted from the amino acid sequence. Due to 11 additional residues in the N-terminus from the His-Tag fragment and enterokinase site, the molecular mass of the N-terminal His-Tag fusion MMLV-RT was approximately 2 kDa larger than without the His-Tag (approximately 56 kDa). According to the BCA assay, the purified MMLV-RT generated the total protein concentration of 0.0084 g L−1. The expression of the enzyme was examined by western blot analysis using HisprobeTM-HRP conjugate. Figure 7 showed that MMLV-RT was detected and had a molecular weight of 58 kDa. The result was in agreement with SDS-PAGE analysis.
Heterologous expression of MMLV-RT in E. coli BL21 star (DE3). A SDS-PAGE analysis using 10% acrylamide gels. Lane M, molecular weight protein marker; Lane 1, crude cell-free extract; Lane 2, purified MMLV-RT protein. B Western blot analysis of MMLV-RT. Lane M, molecular weight protein marker; Lane 1, purified MMLV-RT
3.4 Activity of Recombinant MMLV-RT
In order to confirm the activity of MMLV-RT, an RT-PCR assay was carried out. In the first step of RT-PCR, viral RNA was successfully reverse-transcribed into cDNA by our purified recombinant MMLV-RT. The reagent mixture that consists of random hexamer and oligo d(T) primers, dNTPs, and RNase inhibitor was combined with the commercial (Thermo Fisher Scientific) and purified RTs. The step was continued to PCR amplification using the template obtained in the previous step. The desired result was achieved in the PCR product. The amplification generated a single band with a ~ 400 bp DNA fragment. The recombinant RT was used in different volumes to amplify a 400 bp fragment. The purified recombinant RT showed that it could be used in PCR amplification compared to the commercial RT.
As shown in Fig. 8, the expected amplified DNA is marked in line with an arrow. A 3 µL of respective PCR product was loaded in lanes 1, 2, and 3. The DNA fragment amplified of our recombinant RT showed closely equivalent to that of commercial RT. Negative control was included in RT-PCR assay to detect the possibility of DNA contamination, such as genomic DNA or amplicons of PCR products from previous experiments. Reverse transcription did not occur in this assay because the control contains all the reaction components except for the RT.
A summary of MMLV-RT purification and relevant details regarding total activity, total protein, and specific activity are presented in Table 3. A one-step purification procedure was carried out using Ni-affinity chromatography. In the end, the purification generated 3.59 fold with a 10.74% yield of purified MMLV-RT.
4 Discussion
RT is a well-known enzyme in the biotechnology field due to its numerous applications when combined with PCR including for the expression genes examination, transcript variant detection, and cDNA template synthesis for cloning and sequencing. In general, there are three key activities of RT: (i) the synthesis of the DNA strand complementary to the RNA template by RNA-dependent DNA polymerase, (ii) the degradation of the RNA strand in RNA-DNA hybrids by RNase H endonuclease, and (iii) the conversion of the single-stranded cDNA into double-stranded DNA by DNA-dependent DNA polymerase activity [7, 8]. An earlier study has reported that RT with good catalytic activity, low RNase H activity, and stability at high temperatures are desirable in biotechnological applications [29]. Among various sources of RTs, RT from MMLV is the most frequently and preferably produced in the industry [30]. The current study has attempted to increase the production and performance of the MMLV-RT. Some strategies have been performed to achieve these goals. Firstly, we sought an effective way to produce MMLV-RT using a synthetic gene based on a patent reference in which the original amino acid sequence has been changed at Y139A, T197E, and F139N to diminish the RNase H activity. Several studies have revealed the significant rise of MMLV-RT activity could be reached by mutating the amino acid sequences and generating mutants such as E69K/E302R/W313F/L435G/N454K and L139P/D200N/T330P/L603W/E607K [31, 32].
Secondly, codon optimization was designed to enhance the level of MMLV-RT expression. Although there has been some research on improving enzymatic production, the study of codon optimization for the MMLV-RT-encoding gene is still limited information. When a recombinant enzyme is heterologously expressed, codon optimization is fundamental to perform. Typically, each organism has its own bias and preference to use 61 available codons [33, 34]. In this study, codon optimization of MMLV-RT was performed according to the E. coli codon usage. The coding sequence of MMLV-RT from the original source has been substituted with synonymous codons that encode the same amino acids for E. coli aiming to rise the expression level. The use of the synthetic gene allowed us to perform synonymous codon substitutions in order to obtain optimal codons. Previous studies reported that codon replacement has proven to have a notable impact on gene expression levels and protein folding [35, 36]. The frequency of codons used amongst organisms is diverse and it has a positive correlation with the concentration of tRNA, which can determine the number of amino acids available and the efficiency of protein translation [37, 38]. In other words, codon optimization is essential for producing highly expressed proteins in this study because genes with optimal codons in E. coli are preferable to encode and translate into protein. The presence of rare codons in E. coli tends to decrease the rate of translation, even cause translation error, and resulted in producing truncated recombinant enzymes. All of these things are associated with ribosome stalling. When the tRNA level is depleted, the ribosome can stop the protein synthesis and lead to produce inactive enzymes. Therefore, converting the original DNA sequence into an optimized codon version is vital to improve translation efficiency, which impacts protein conformation and stability [39, 40, 41]
Furthermore, one of the primary indexes used for codon optimization to predict the protein expression level is the Codon Adaptation Index (CAI). The CAI is a simple and effective measure of synonymous codon usage bias [42]. CAI value reflects how well our synthetic gene sequence can adapt to the new expression host. CAI values range from 0 to 1, indicating the less frequently used codons to only the most abundant used codons. However, the optimization approach using ‘one amino acid = one codon’ or ‘CAI = 1’ has several short-coming. According to Villalobos et al., using one codon in highly expressed protein could make an imbalanced tRNA pool resulting in tRNA depletion and increased frameshift. Moreover, we cannot avoid repetitive elements and mRNA secondary structure in DNA sequence, which can harm the protein synthesis [43]. In this study, we used the ‘guided random’ method to vary the use of codons and removed the rare codons. The result of codon optimization demonstrated that the CAI value of the final codon-optimized sequence was 0.789 out of 1. It indicates that the codons used in our gene are mostly for tRNA abundant in cells, assuming it would produce a highly expressed gene.
The mRNA folding near TIR was also known as the crucial effect on translation efficiency. A previous study showed that increasing the mRNA folding energy near the ribosome binding site has been impactful to improve the expression level of recombinant GCSF protein in the E. coli system [44]. Moreover, another related literature has revealed that the unstable mRNA at the TIR could facilitate the efficient recognition of the start codon [45]. Hence, we carried out synonymous codon substitution at the 5’-terminal end of the initial sequence. The result showed that the mRNA folding energy value of the final sequence was increased by more than 50% from the initial sequence and indicated the more efficient translation initiation process.
Amongst the bacteria, E. coli is the most popular system used to produce recombinant proteins. This host is favored for its ease of growing in an inexpensive medium. However, there are major challenges faced by employing E. coli expression system, including the expression of complex proteins with many rare codons or disulfide bonds and toxic proteins [46, 47]. In this experiment, we initially analyzed the MMLV-RT protein characteristics to find out the suitability of E. coli as the expression host. By using SOLUPROT v.1 and DISULFIND software we found that MMLV-RT could be expressed in the E. coli system which was indicated by high solubility value and the absence of disulfide bonds. This is crucial to comprehend since the protein consists of disulfide bonds in nature and tends to form inclusion bodies (IBs) when it is expressed in the E. coli host [33].
Lastly, the optimization of culture conditions was conducted in the effort to find the optimum conditions for improved protein expression. The production of recombinant enzymes in the E. coli system, despite its potential to be applied for obtaining the desired enzyme, poses several hurdles. Protein aggregation such as IBs becomes a common problem that should be addressed. Even after some precautions have been taken such as performing an initial assessment of suitability for expression in E. coli and the analysis of MMLV-RT solubility, the formation of IBs still could be discovered. The formation of IBs may be due to an unbalanced equilibrium among proper folding, aggregation, and degradation, which can be triggered by the high rate of protein expression. For that reason, the temperature condition in the post-induction phase was modified by lowering it to 18 °C. Decreasing the culture temperature may result in the low rate of protein expression and prevent the potential misfolding of the enzymes due to the high rate of protein expression [48]. Moreover, the expression of insoluble proteins correlated with the higher temperatures and lower amount of time [49]. In our study, the culture was grown at 37 °C to obtain high cell density, then incubated overnight at 18 and 27 °C. Employing induction temperature at 27 °C did increase the protein yield. However, further enhancement was obtained by shifting the post-induction temperature lower to 18 °C. The strategy of shifting the culture to the lower temperature has been applied as well in other studies [50, 51]. Furthermore, the present study bears out the result found by Chen et al. for being successful to obtain MMLV-RT in E. coli via culturing at 28 °C [52]. Our finding found that the shifting apparently did not affect the bacterial biomass at the end of cultivation, as we can observe that the cultures were harvested at similar OD600 value. Other strategies are available to minimize the formation of IBs. To conform the expression conditions, performing expression host engineering via co-expressing chaperone could also be employed, among others [48]. Nevertheless, our study succeeded in over-expressing the MMLV-RT as well as improving its solubility by means of shifting to the lower post-induction time.
After obtaining the suitable post-induction temperature, we opted to vary the IPTG concentration. Under various IPTG concentrations employed in this study, the protein yield obtained seemed to be indifferent. In the current study, IPTG of 0.1 mM is sufficient to induce the expression of the desired enzyme. Previously, similar studies have used higher IPTG concentration (0.6 mM) to induce the production of MMLV-RT. They found that lowering the temperature was not adequate to increase the solubility of the desired enzyme. Upon employing lower temperature and IPTG concentration, they succeeded in obtaining the improvement of solubility [50, 52]. The lower IPTG concentration, combined with lower post-incubation temperature was supposed to be beneficial for the production of MMLV-RT in this study. Fazaeli et al. also reported that to obtain the highest yield of recombinant cholesterol oxidase, they only required the induction of low IPTG concentration at 0.1 mM. Improving the IPTG concentration did not linearly correlated with the protein yield obtained [53]. In fact, the high concentration of inducer has probably impacted on metabolic burden instead of improving the target protein [54, 55].
Further optimization was performed by varying post-induction time. In this study, we found that prolonging incubation time after induction until 24 h led to the highest levels of target protein concentration. A similar strategy was also applied by Sina et al. to obtain the highest level of soluble recombinant GST-hD2 by employing low cultivation temperature in the presence of low concentrations of IPTG under long incubation time [51]. In general, the strategy to combine the optimization of codon and culture conditions was successful to generate the highly improved expression of MMLV-RT.
Protein expression of MMLV-RT in bacterial cells was examined by SDS-PAGE and Western blotting. Samples obtained from each treatment and purification step were loaded on a 10% polyacrylamide gel to assess and evaluate the purity, solubility, and yield. Compared with other related studies, our study showed positive results in terms of total purified protein of reverse transcriptase obtained which was able to reach 8.4 mg L−1 using HisTrap™ HP column for purification, while another data obtained by Lu et al. has achieved at 0.075 mg L−1 using RNHI affinity column [56]. In addition, data reported in another literature has also revealed the lower total protein (3.8 mg L−1) using Q-sepharose column than our data [57]. It indicates that our study can be potential and prospective to be further developed.
5 Conclusions
The recombinant MMLV-RT was successfully overexpressed in E. coli BL21 star (DE3) under optimized conditions: initial pre-induction OD600 at 0.4 with 0.1 mM IPTG at 18 °C of post-induction temperature and 24 h of post-induction time. Protein concentration could be increased up to 85-fold after optimizing the culture conditions. In this preliminary study, the purified MMLV-RT generated 5275.02 mg U−1 of specific activity with 3.59-fold purification and 10.74% of yield. It exhibited the potent activity to be applied in RT-PCR assay. This study provides fruitful strategies to enhance the recombinant enzyme of MMLV-RT in both production and performance. The enzyme can be potentially used and promising to reverse-transcribe the viral RNA into cDNA. Moreover, further studies are needed to characterize the mutant MMLV-RT in this study in comparison to the native one.
Data Availability
The authors declare that all data supporting the findings of this study are available within the article.
References
Baltimore D (1970) RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226:1209–1211. doi:https://doi.org/10.1038/2261209a0
Temin HM, Mizutani S (1970) RNA-dependent DNA polymerase in virions of rous sarcoma virus. Nature 226:1211–1213. https://doi.org/10.1038/2261211a0
Mizutani S, Boettiger D, Temin HM (1970) A DNA-dependent DNA polymerase and a DNA endonuclease in virions of rous sarcoma virus. Nature 228:424–427
Zajac P, Islam S, Hochgerner H, Lönnerberg P, Linnarsson S (2013) Base preferences in non-templated nucleotide incorporation by MMLV derived reverse transcriptases. PLoS ONE. https://doi.org/10.1371/journal.pone.0085270
Hu WS, Hughes SH (2012) HIV-1 reverse transcription. Cold Spring Harb Perspect Med. https://doi.org/10.1101/cshperspect.a006882
Tian L, Kim MS, Li H, Wang J, Yang W (2018) Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid. Proc Natl Acad Sci USA 115:507–512. doi:https://doi.org/10.1073/pnas.1719746115
Coffin JM, Fan H (2016) The discovery of reverse transcriptase. Annu Rev Virol 3:29–51. https://doi.org/10.1146/annurev-virology-110615-035556
Costa C, Giménez-Capitán A, Karachaliou N, Rosell R (2013) Comprehensive molecular screening: from the RT-PCR to the RNA-seq. Transl Lung Cancer Res 2:87–91. doi: https://doi.org/10.3978/j.issn.2218-6751.2013.02.05
Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, Sasso M, Gregg AC, Soares DJ, Beskid TR, Jervey SR, Liu C (2020) Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci 6(5):591–605. doi:https://doi.org/10.1021/acscentsci.0c00501
Goudouris ES (2021) Laboratory diagnosis of COVID-19. Jornal de pediatria 97:7–12. https://doi.org/10.1016/j.jped.2020.08.001
Nishimura K, Yokokawa K, Hisayoshi T, Fukatsu K, Kuze I, Konishi A, Mikami B, Kojima K, Yasukawa K (2015) Preparation and characterization of the RNase H domain of moloney murine leukemia virus reverse transcriptase. Protein Expr Purif 113:44–50. doi: https://doi.org/10.1016/j.pep.2015.04.012
Konishi A, Ma X, Yasukawa K (2014) Stabilization of moloney murine leukemia virus reverse transcriptase by site-directed mutagenesis of the surface residue Val433. Biosci Biotechnol Biochem 78:147–150
Mizuno M, Yasukawa K, Inouye K (2010) Insight into the mechanism of the stabilization of moloney murine leukemia virus reverse transcriptase by eliminating RNase H activity. Biosci Biotechnol Biochem 74:440–442
Marintcheva B (2017) Harnessing the power of viruses. Academic Press, London
Katano Y, Hisayoshi T, Kuze I, Okano H, Ito M, Nishigaki K, Takita T, Yasukawa K (2016) Expression of moloney murine leukemia virus reverse transcriptase in a cell-free protein expression system. Biotechnol Lett 38:1203–1211. doi:https://doi.org/10.1007/s10529-016-2097-0
Potter K, Rosenthal K (2013) High fidelity of reverse transcriptases and the uses thereof (US Patent No. 8,541,219 B2). https://patents.google.com/patent/US7056716B2/en
Hon J, Marusiak M, Martinek T, Kunka A, Zendulka J, Bednar D, Damborsky J (2021) SoluProt: prediction of soluble protein expression in Escherichia coli. Bioinformatics 37:23–28. doi:https://doi.org/10.1093/bioinformatics/btaa1102
Ceroni A, Passerini A, Vullo A, Frasconi P (2006) DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res 34:177. https://doi.org/10.1093/nar/gkl266
Puigbò P, Bravo IG, Garcia-Vallve S (2008) CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct 3:38. doi:https://doi.org/10.1186/1745-6150-3-38
Reuter JS, Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform 11:129. https://doi.org/10.1186/1471-2105-11-129
Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, McEntaggart NO, Sailor CA, Dawson RB, Peek AS (2008) IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res 36:163–169. doi: 10.1093nar/gkn198
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. https://doi.org/10.1093/nar/gkg563
Sievers F, Higgins DG (2014) Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079:105–116. doi:https://doi.org/10.1007/978-1-62703-646-7_6
Chung CT, Niemela SL, Miller RH (1989) One step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA 86(7):2172–2175
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Larentis AL, Fabiana J, Quintal M, Esteves S, Vareschini DT, Vicente F, Almeida RD (2014) Evaluation of pre-induction temperature, cell growth at induction and IPTG concentration on the expression of a leptospiral protein in E. coli using shaking flasks and microbioreactor. BMC Res Notes 7:671. doi: https://doi.org/10.1186/1756-0500-7-671
Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85
Quick J (2020) nCoV-2019 sequencing protocol v3 (LoCost) V3. Available from: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye
Martín-Alonso S, Frutos-Beltrán E, Menéndez-Arias L (2021) Reverse transcriptase: from transcriptomics to genome editing. Trends Biotechnol 39:194–210
Wulf MG, Maguire S, Humbert P, Dai N, Bei Y, Nichols NM, Corrêa IR, Guan S (2019) Non-templated addition and template switching by moloney murine leukemia virus (MMLV)-based reverse transcriptases co-occur and compete with each other. J Biol Chem 294:18220–18231. doi: https://doi.org/10.1074/jbc.RA119.010676
Arezi B, Hogrefe H (2009) Novel mutations in moloney murine leukemia virus reverse transcriptase increase thermostability through tighter binding to template-primer. Nucleic Acids Res 37:473–481
Baranauskas A, Paliksa S, Alzbutas G (2012) Generation and characterization of new highly thermostable and processive M-MuLV reverse transcriptase variants. Protein Eng Des Sel 25:657–668
Kaur J, Kumar A, Kaur J (2018) Strategies for optimization of heterologous protein expression in E. coli: roadblocks and reinforcements. Int J Biol Macromol 106:803–822. doi: https://doi.org/10.1016/j.ijbiomac.2017.08.080
Inouye S, Sahara-Miura Y, Sato JI, Suzuki T (2015) Codon optimization of genes for efficient protein expression in mammalian cells by selection of only preferred human codons. Protein Expr Purif 109:47–54
Elena C, Ravasi P, Castelli ME, Peirú S, Menzella HG (2014) Expression of codon optimized genes in microbial systems: current industrial applications and perspectives. Front Microbiol 5:21. doi:https://doi.org/10.3389/fmicb.2014. 00021
Menzella HG (2011) Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli. Microb Cell Fact 10:15. doi:https://doi.org/10.1186/1475-2859-10-15
Gouy M, Gautier C (1982) Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res 10:7055–7074
Tokuoka M, Tanaka M, Ono K, Takagi S, Shintani T, Gomi K (2008) Codon optimization increases steady-state mRNA levels in Aspergillus oryzae heterologous gene expression. Appl Environ Microbiol 74:6538–6546. doi: https://doi.org/10.1128/AEM.01354-08
Ikemura T (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol 151:389–409
Gaspar P, Oliveira JL, Frommlet J, Santos MA, Moura G (2012) EuGene: maximizing synthetic gene design for heterologous expression. Bioinformatics 28:683–2684
Al-Hawash AB, Zhang X, Ma F (2017) Strategies of codon optimization for high-level heterologous protein expression in microbial expression systems. Gene Rep 9:46–53
Fu H, Liang Y, Zhong X, Pan Z, Huang L, Zhang H, Xu Y, Zhou W, Liu Z (2020) Codon optimization with deep learning to enhance protein expression. Sci Rep 10:17617. https://doi.org/10.1038/s41598-020-74091-z
Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S (2016) Gene designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinform 7:1–8. https://doi.org/10.1186/1471-2105-7-285
Dewi KS, Fuad AM (2020) Improving the expression of human granulocyte colony stimulating factor in Escherichia coli by reducing the GC-content and increasing mRNA free folding energy at 5’-terminal end. Adv Pharm Bull 10:610–616. https://doi.org/10.34172/apb.2020.073
Gu W, Zhou T, Wilke CO (2010) A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1000664
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172. https://doi.org/10.3389/fmicb.2014.00172
Ahmad M, Hirz M, Pichler H, Schwab H (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98:5301–5317. https://doi.org/10.1007/s00253-014-5732-5
Bhatwa A, Wang W, Hassan YI, Abraham N, Li X (2021) Challenges associated with the formation of recombinant protein inclusion bodies in Escherichia coli and strategies to address them for industrial applications. Front Bioeng Biotechnol 9:1–18
Gutiérrez-gonzález M, Farías C, Tello S, Pérez-etcheverry D, Romero A, Zúñiga R, Ribeiro CH (2019) Optimization of culture conditions for the expression of three different insoluble proteins in Escherichia coli. Sci Rep 9:16850. doi: https://doi.org/10.1038/s41598-019-53200-7
Jhamb K, Sahoo DK (2012) Production of soluble recombinant proteins in Escherichia coli : effects of process conditions and chaperone co-expression on cell growth and production of xylanase. Bioresour Technol 123:135–143. doi: https://doi.org/10.1016/j.biortech.2012.07.011
Sina M, Farajzadeh D, Dastmalchi S (2015) Effects of environmental factors on soluble expression of a humanized anti-TNF-α scFv antibody in Escherichia coli. Adv Pharm Bull 5:455–461. doi: https://doi.org/10.15171/apb.2015.062
Chen Y, Xu AW, Sun AQ (2009) A novel and simple method for high-level production of reverse transcriptase from moloney murine leukemia virus (MMLV-RT) in Escherichia coli. Biotechnol Lett 31:1051–1057
Fazaeli A, Golestani A, Lakzaei M, Sadat S, Varaei R, Id MA (2019) Expression optimization, purification, and functional characterization of cholesterol oxidase from Chromobacterium sp. DS1. PLoS ONE 20114:1–15. https://doi.org/10.1371/journal.pone.0212217
Donovan RS, Robinson CW, Glick BR (1996) Review: optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J Ind Microbiol 16:145–154. doi: https://doi.org/10.1007/BF01569997
Glick BR (1995) Metabolic load and heterologous gene expression. Biotechnol Adv 13:247–261. doi: https://doi.org/10.1016/0734-9750(95)00004-a
Lu M, Ngo W, Mei Y, Munshi V, Burlein C, Loughran MH, Williams PD, Hazuda DJ, Miller MD, Grobler JA, Diamond TL, Lai MT (2010) Purification of untagged HIV-1 reverse transcriptase by affinity chromatography. Protein Expr Purif 71:231–239. doi:https://doi.org/10.1016/j.pep.2010.01.001
Silprasit K, Thammaporn R, Hannongbua S, Choowongkomon K (2008) Cloning, expression, purification, determining activity of recombinant HIV-1 reverse transcriptase. Nat Sci 42:231–239
Acknowledgements
This work was supported by the research grant from Indonesia Endowment Fund for Education (Grant No: KEP-63/LPDP/2020). We acknowledge the COVID-19 diagnostic testing team and Genomic Surveillance of SARS-CoV-2 (VENOMCoV) team of Biosafety Level 3 (BSL-3) Laboratory, National Research and Innovation Agency (BRIN)-Indonesia for providing viral RNA samples to this study.
Author information
Authors and Affiliations
Contributions
IN, FAL, EA, and KSD contributed equally to this study. All authors were involved in the research conception and design. Materials preparation, data collection, and interpretation, as well as manuscript writing, were performed by IN, FAL, EA, and KSD Editing was conducted by AA and AT This study was supervised by WK and PL. All authors read the revised manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nuryana, I., Laksmi, F.A., Agustriana, E. et al. Expression of Codon-Optimized Gene Encoding Murine Moloney Leukemia Virus Reverse Transcriptase in Escherichia coli. Protein J 41, 515–526 (2022). https://doi.org/10.1007/s10930-022-10066-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-022-10066-5