Abstract
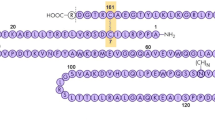
Determination of biological activity and its comparison with clinical behavior is important in the quality assessment of therapeutic glycoproteins. In vivo studies are usually employed for evaluating bioactivity of these glycomolecules. However, alternative methods are required to simplify the bioassay and avoid ethical issues associated with in vivo studies. Negatively charged sialic acid residues are known to be critical for in vivo bioactivity of rHuEPO. To address this need, we employed the human acute myeloid leukemia cell line UT-7 for the determination of proliferative stimulation induced by rHuEPO. Relative potencies of various intact and sugar-trimmed rHuEPO preparations were estimated using the International Standard for Human r-DNA derived EPO (87/684) as a reference for bioactivity. The cellular response was measured with a multi-channel photometer using a colorimetric microassay, based on the metabolism of the Resazurin sodium by cell viability. For a resourceful probing of physiological features of rHuEPO with significance, we obtained partly or completely desialylated rHuEPO digested by the neuraminidase enzyme without degradation of carbohydrates. Two-fold higher specific activity was shown by asialoerythropoietin in in vitro analysis compared with the sialoerythropoietin. Further, computational studies were also carried out to construct the 3D model of the erythropoietin (EPO) protein structure using standard comparative modeling methods. The quality of the model was validated using Procheck and protein structure analysis (ProSA) server tools. N–glycan units were constructed; moreover, EPO protein was glycosylated at potential glycosylation amino acid residue sites. The method described should be suitable for potency assessments of pharmaceutical formulations of rHuEPO (European Pharmacopeia, 2016).












Similar content being viewed by others
Abbreviations
- rHuEPO:
-
Recombinant Human Erythropoietin
- EPO:
-
Erythropoietin
- G-CSF:
-
Granulocyte Colony Stimulating Factor
- GM-CSF:
-
Granulocyte Macrophage Colony Stimulating Factor
- IL-3:
-
Interleukin 3
- IL-9:
-
Interleukin 9
- IGF-1:
-
Insulin Like Growth Factor 1
- FBS:
-
Fetal Bovine Serum
- BFU-E:
-
Burst Forming Unit-Erythroid
- CFU-E:
-
Colony Forming Unit-Erythroid
- CFU-GEMM:
-
Colony Forming Unit-Granulocyte, Erythroid, Macrophage, Megakaryocyte
- Erythropoietin Receptor:
-
EPO-R
- Chinese Hamster Overy:
-
CHO
- MTS:
-
(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium).
References
Erythropoietin concentrate solution, Assay, European Pharmacopeia (2016)
Molineux G, Sinclair A (2009) Biology of erythropoietin. In: Parnham MJ, Bruinvels J, eds. Erythropoietins, erythropoietic factors, and erythropoiesis (2nd ed.) Basel: Birkhauser 2009:41–60
Wang FF, Kung CKH, Goldwasser E (1985) Some chemical properties of human erythropoietin. Endocrinology 116:2286–2292
Roberts D, Smith DJ (1994) Erythropoietin: Induction of synthesis to signal transduction. J Mol Endocrinol 12:131–148
Choi D, Kim M, Park J (1996) Erythropoietin: physico and biochemical analysis. J Chromatogr B Biomed Appl 687:189–199
Dordal MS, Wang FF, Goldwasser E (1985) The role of carbohydrate in erythropoietin action. Endocrinology 116:2293–2299
Lai PH, Everett R, Wang FF (1986) Structural characterization of human erythropoietin. J Biol Chem 261:3116–3121
Tran AD, Park S, Lisi PJ, Huynh OT, Ryall RR, Lane PA (1991) Separation of carbohydrate-mediated microheterogeneity of recombinant human erythropoietin by free solution capillary electrophoresis. Effects of pH, buffer type and organic additives. J Chromatogr A 542:459–471
Rudd PM, Dwek RA (1997) Glycosylation: heterogeneity and the 3D structure of proteins. Crit Rev Biochem Mol Biol 32(1):1–100
Sasaki H, Bothner B, Dell A, Fukuda M (1987) Carbohydrate structure of erythropoietin expressed in Chinese hamster ovary cells by a human erythropoietin cDNA. J Biol Chem 262(25):12059–12076
Rush RS, Derby PL, Smith DM, Merry C, Rogers G, Rohde MF, Katta V (1995) Microheterogeneity of erythropoietin carbohydrate structure. Anal Chem 67(8):1442–1452
Kiss Z, Elliott S, Jedynasty K, Tesar V, Szegedi J (2010) Discovery and basic pharmacology of erythropoiesisstimulating agents (ESAs), including the hyperglycosylated ESA, darbepoetin alfa: an update of the rationale and clinical impact. Eur J Clin Pharmacol 66(4):331–340
Miura Y, Komatsu N, Suda T (1990) Growth and differentiation of two human megakaryoblastic cell lines: CMK and UT-7. Prog Clin Biol Res 356:259
Komatsu N, Fujita H (1993) Induced megakaryocytic maturation of the human leukemic cell line, UT-7, results in down-modulation of erythropoietin receptor gene expression. Cancer Res 53(5):1156–1161
Mire-Sluis AR, Gaines-Das R, Thorpe R (1995) Immunoassays for detecting cytokines: what are they really measuring? J Immunol Methods 186:157–160
Bird C, Wadhwa M, Thorpe R (1991) Development of immunoassays for human interleukin 3 and interleukin 4, some of which discriminate between different recombinant DNA-derived molecules. Cytokine 3:562–567
Moller B, Mogensen SC, Wendelboe P, Bendtzen K, Petersen CM (1991) Bioactive and inactive forms of tumor necrosis factor- in spinal fluid from patients with meningitis. J Infect Dis 163:886–889
Onyango JD, Burri C, Brun R (2000) An automated biological assay to determine levels of the trypanocidal drug melarsoprol in biological fluids. Acta Trop 74:95–100
Gazzano-Santoro H, Chen A, Casto B, Chu H, Gilkerson E, Mukku V, Canova-Davis E, Kotts C (1999) Validation of a rat pheochromocytoma (PC12)-based cell survival assay for determining biological potency of recombinant human nerve growth factor. J Pharm Biomed Anal 21:945–959
Canosi U, Mascia M, Gazza L, Serlupi-Crescenzi O, Donini S, Antonetti F, Galli G (1996) A highly precise reporter gene bioassay for type I interferon. J Immunol Methods 199(1):69–76
Adams RL (1969) The effect of endogenous pools of thymidylate on the apparent rate of DNA synthesis. Exp Cell Res 56:55–58
Khoo SK, Hurst T, Webb MJ, Dickie G, Kearsley J, Parsons PG, Mackay EV (1988) Measurement of tumor cell activity in short-term primary culture. Clinical significance in women with ovarian cancer. Cancer 61:1579–1586
Westermark K, Karlsson FA, Westermark B (1983) Epidermal growth factor modulates thyroid growth and function in culture. Endocrinology 112(5):1680–1686
Mosmann T (1983) Epidermal growth factor modulates thyroid growth and function in culture. J Immunol Methods 65:55–63
Raz B, Iten M, Grether-Buhler Y, Kaminsky R, Brun R (1997) The Alamar Blue® assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop 68:139–147
Crouch SPM, Kozlowski R, Slater KJ, Fletcher J (1993) The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J Immunol Methods 160:81–88
Metta MK, Kunaparaju RK, Tantravahi S (2016) Rapid amplification system for recombinant protein production in Chinese Hamster Ovary (CHO) Cells. Cell Mol Biol 62(2):101–106
Erythropoietin concentrate solution (2016) Electrophoresis 2.2.31, European Pharmacopeia
Kreft S, Kreft M (2009) Quantification of dichromatism: a characteristic of color in transparent materials. J Opt Soc Am A 26:1576–1581
Page B, Page M, Noel C (1993) A new fluorimetric assay for cytotoxicity measurements in vitro. Int J Oncol 3:473–476
Matsumoto K, Yamada Y, Takahashi M, Todoroki T, Mizoguchi K, Misaki H, Yuki H (1990) Fluorometric determination of carnitine in serum with immobilized carnitine dehydrogenase and diaphorase. J Clin Chem 36:2072–2076
Belinsky M, Jaiswal AK (1993) NAD(P)H: quinone oxidoreductase1DT-diaphorase expression in normal and tumor tissues. Cancer Metastasis Rev 12:103–117
Chikuba K, Yubisui T, Shirabe K, Takeshita M (1994) Cloning and nucleotide sequence of a cDNA of the human erythrocyte NADPH-flavin reductase. Biochem Biophys Res Commun 198:1170–1176
Moon TW, Mommsen TP (2005) Biochemistry and molecular biology of fishes. Environ Toxicol 6:51–56
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36(Web Server issue):W5-9
Drozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucl Acids Res 43(W1):W389–W394
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucl Acids Res 28(1):235–242
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Webb B, Sali A (2014) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 47:5.6.1–32
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815
Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6(5):1509–1519
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three imensional structures of proteins. Nucleic Acids Res 35(suppl 2):W407–W410
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17(4):355–362
Bohne-Lang A, von der Lieth CW (2005) GlyProt: in silico glycosylation of proteins. Nucleic Acids Res 33(suppl 2):W214–W219
Bohne A, Lang E, von der Lieth CW (1999) SWEET- WWW-based rapid 3D construction of oligo- and polysaccharides. Bioinformatics 15(9):767–768
Bohne A, Lang E, von der Lieth CW (1999) W3-SWEET: carbohydrate modeling by internet. Mol Model Annu 4(1):33–43
Fukuda MN, Sasaki H, Lopez L, Fukuda M (1989) Survival of recombinant erythropoietin in the circulation: the role of carbohydrates. Blood 73:84–89
Spivak JL, Hogans BB (1989) The in vivo metabolism of recombinant human erythropoietin in the rat. Blood 73:90–99
Tsodikov OV, Record MT, Sergeev YV (2002) Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem 23(6):600–609
Moore E, Bellomo R (2011) Erythropoietin (EPO) in acute kidney injury. Ann Intensive Care 1:3:53
Tanaka T, Nangaku M (2012) Recent advances and clinical application of erythropoietin and erythropoiesis-stimulating agents. Exp Cell Res 318(9):1068–1073
Elliott S, Egrie J, Browne J, Lorenzini T, Busse L, Rogers N, Ponting I (2004) Control of rHuEPO biological activity: the role of carbohydrate. Exp Hematol 32:1146–1155
Takeuchi M, Kobata A (1991) Structures and functional roles of the sugar chains of human erythropoietins. Glycobiology 1:337–346
Ramos AS, Schmidt CA, Andrade SS, Fronza M, Rafferty B, Dalmora SL (2003) Biological evaluation of recombinant human erythropoietin in pharmaceutical products. Braz J Med Biol Res 36(11):1561–1569
Sinclair AM (2013) Erythropoiesis stimulating agents: approaches to modulate activity. Biologics 7:161–174
Acknowledgements
I am grateful to Usha Biotech and GITAM University for their support in carrying out this research study. I would also like to acknowledge the Head, Department of Chemistry and the Principal, University College of Science, Osmania University for providing the facilities to carry out this bioinformatics study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Metta, M., Malkhed, V., Tantravahi, S. et al. Development of an in vitro Bioassay for Recombinant Human Erythropoietin (rHuEPO) Based on Proliferative Stimulation of an Erythroid Cell Line and Analysis of Sialic Acid Dependent Microheterogeneity: UT-7 Cell Bioassay. Protein J 36, 112–122 (2017). https://doi.org/10.1007/s10930-017-9704-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-017-9704-3