Abstract
Salicortin is a phenolic glucoside produced in Salicaceae as a chemical defense against herbivory. The specialist lepidopteran herbivorous larvae of Cerura vinula are able to overcome this defense. We examined the main frass constituents of C. vinula fed on Populus nigra leaves, and identified 11 quinic acid derivatives with benzoate and/or salicylate substitution. We asked whether the compounds are a result of salicortin breakdown and sought answers by carrying out feeding experiments with highly 13C-enriched salicortin. Using HRMS and NMR analyses, we were able to confirm that salicortin metabolism in C. vinula proceeds through deglucosylation and ester hydrolysis, after which saligenin is oxidatively transformed into salicylic acid and, eventually, conjugated to quinic acid. To the best of our knowledge, this is the first report of a detoxification pathway based on conjugation with quinic acid.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salicinoids – glycosides derived from salicyl alcohol (saligenin) – are defensive chemicals of Populus species and of other members of the Salicaceae family (Lindroth 1991; Palo 1984). The structural diversity of salicinoids arises from their modular composition, which comprises a saligenin core unit, a glucose moiety and an organic acid. The most representative salicinoid in Populus is salicortin (12), as so far it has been found in all investigated species (Boeckler et al. 2011; Thieme 1964). Numerous studies have investigated the effect of salicinoids on lepidopteran herbivorous insects, such as Papilio glaucus (Lindroth 1991), Choristoneura conflictica (Clausen et al. 1989), Malacosoma disstria and Lymantria dispar (Lindroth and Hemming 1990) or Operophtera brumata (Boeckler et al. 2016; Ruuhola et al. 2001). Coleopteran larvae belonging to the Chrysomelidae family can sequester salicinoids and use their host plant’s chemical defense to ward off predators (Burse et al. 2009). Recently, a comprehensive study of the generalist herbivore L. dispar (Boeckler et al. 2016) provided the first detailed description of how a lepidopteran detoxifies salicinoid compounds. The digestive degradation of salicinoids in the insect gut (Haruta et al. 2001; Lindroth 1988; Ruuhola et al. 2003) results in saligenin and an o-quinone (Clausen et al. 1989; Julkunen-Tiitto and Meier 1992; Knuth et al. 2011). Both metabolites are known for their toxic and feeding-deterrent activities (Boeckler et al. 2011; Clausen et al. 1989; Ruuhola et al. 2001).
The larva of the lepidopteran Cerura vinula is native to Europe and Asia. In the temperate climate zone of Central Europe, imagines appear from April to August. Females lay eggs on branches and leaves of their host plants. As the insects’ nutritional spectrum is limited to plants of the Salicaceae family (Salix and Populus species), they are regarded as specialist herbivores (Ali and Agrawal 2012; Hintze-Podufal 1970). Numerous metabolic studies (recent examples: Beran et al. 2014; Joussen et al. 2012; Shelomi et al. 2016) have addressed the question whether a specialist herbivore uses a distinct mechanism to cope with the defense of its host plant. In the present publication, we used a double-track strategy: the careful structural identification of metabolic products arising from the diet of the specialist herbivore resulted in an assumption about how the main chemical defense compounds were transformed during digestion. This hypothesis was then supported using stable isotope labeling which eventually allowed us to develop a new salicinoid degradation pathway.
Methods and Materials
General Information
In order to address the question of how C. vinula is able to deactivate the salicinoid defense of its host plant, several experimental approaches were employed. It was necessary to develop UPLC-MS and NMR protocols for the identification of the main metabolites as well as an HPLC-SPE protocol for the degradation-free workup of the compounds of interest. [13C]-Labeling of salicinoids was accomplished in planta by growing plants under a [13C]CO2-enriched atmosphere in a dedicated growth chamber and subsequent isolation of [U-13C]salicortin from leaf material. Experimental details and procedures can be found in the following section.
NMR spectra for the structure elucidation of acylated quinic acids 1 to 11 were recorded on a Bruker Avance III HD 700 MHz spectrometer, equipped with a 1.7 mm TCI microcryoprobe (Bruker Biospin, Rheinstetten, Germany) using NMR tubes of 1.7 mm outer diameter. NMR spectra of frass extracts from C. vinula larvae after [U-13C]salicortin feeding experiments were obtained on a Bruker Avance III HD 500 MHz NMR spectrometer equipped with a 5 mm TCI cryoprobe (Bruker Biospin) using NMR tubes of 5 mm outer diameter. NMR spectra for the characterization of in vivo-generated [U-13C]salicortin were recorded on a Bruker Avance III HD 400 MHz NMR spectrometer equipped with a 5 mm BBFO probe (Bruker Biospin) using NMR tubes of 5 mm outer diameter. NMR spectra were recorded using MeOH-d 4 as a solvent. Chemical shifts were referenced to the residual solvent peaks at δH 3.31 and δC 49.15. Data acquisition and processing were accomplished using TopSpin 3.2. Standard pulse programs as implemented in TopSpin were used for data acquisition.
The ultra-high-performance liquid chromatography–electrospray ionization–tandem mass spectrometry system (UHPLC–ESI–MS/MS) for structure elucidation and analysis of frass compounds after [U-13C]salicortin labeling consisted of an Ultimate 3000 series RSLC (Dionex, Sunnyvale, CA, USA) and a Q Exactive Plus - Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) using heated-electrospray ionization (H-ESI). H-ESI source parameters were set to 4 kV for spray voltage and 35 V for transfer capillary voltage at a capillary temperature of 300 °C. The samples were measured in positive and negative ionization mode in the mass range of m/z 100 to 1000 using 70,000 m/Δm resolving power in the Orbitrap mass analyzer.
UHPLC-ESI-MS/MS for hemolymph analysis was performed with an Ultimate 3000 series RSLC (Dionex) and LTQ - Orbitrap XL mass spectrometer (Thermo Fisher Scientific) in which ionization was accomplished using electrospray ionization (ESI). ESI source parameters were set to 4 kV for spray voltage, 35 V for transfer capillary voltage at a capillary temperature 275 °C. The samples were measured in negative ionization mode in the mass range of m/z 100 to 1000 using 30,000 m/Δm resolving power in the Orbitrap mass analyzer. All UHPLC systems used an Acclaim C18 column (150 × 2.1 mm, 2.2 μm, Dionex, Sunnyvale, CA, USA) for chromatographic separation. HRMS data were evaluated and interpreted using Xcalibur software (Thermo Fisher Scientific, Waltham, MA, USA).
HPLC-ESI-MS of acylated quinic acids 1–11 was performed on an Agilent 1100 HPLC system, consisting of a degasser, quaternary solvent delivery pump G1311A, an autosampler G1313A (Agilent Technologies, Waldbronn, Germany), a photodiode array detector (detection 200–700 nm; J&M Analytik, Aalen, Germany) and an Esquire 3000 ion trap mass spectrometer (Bruker Daltonik, Bremen, Germany). The column outlet was connected to a Bruker/Spark Holland Prospect 2 solid-phase extraction (SPE) system (Bruker Biospin) for post-column SPE trapping on HySphere resin GP cartridges. To reduce the eluotropic capacity of the HPLC solvent mixture, water was added with a flow rate of 2.5 ml min−1 using a make-up pump (Knauer, Berlin, Germany).
[U-13C]Salicortin was chromatographically purified on an Agilent 1100 HPLC system, consisting of a degasser G1322A, a binary pump G1312A, an autosampler G1313A and a photodiode array detector G1315B (Agilent Technologies). The column outlet was connected to an Advantec CHF122SB fraction collector (Jasco, Gross-Umstadt, Germany) triggered by a relay board from the Agilent 1100. HPLC separations were carried out using an Isis RP-18e column (250 × 4.6 mm, 5 μm particle size) (Macherey-Nagel, Düren, Germany). Solvents were evaporated with a rotary evaporator Rotavapor R-114 (Büchi Labortechnik, Flawil, Switzerland) and a Genevac HT-4X vacuum centrifuge (Genevac, Ipswich, UK). Homogenization was carried out with a MINILYS cell disruptor (Bertin Technologies, Montigny-le-Bretonneux, France). Solvents used for extraction and chromatographic separation were purchased from Carl Roth (Karlsruhe, Germany) and VWR International (Darmstadt, Germany), and used without further purification. Acetonitrile and water (hypergrade for LCMS) used for UHPLC-ESI-MS/MS were purchased from Merck (Darmstadt, Germany), and formic acid (eluent additive for LC-MS) was obtained from Sigma Aldrich (Steinheim, Germany). Water used for HPLC was obtained from a Milli-Q Synthesis A 10 purifier (Merck). 13CO2 (isotopic purity 99 atom% 13C, <3 atom% 18O) and Phytacon™ vessels (H 140 mm, base diam. 86 mm) used as arenas for the larvae feeding experiments were purchased from Sigma-Aldrich (Taufkirchen, Germany). HR-X SPE cartridges (500 mg sorbent/6 ml volume), folded filters (90 mm) and paper filters (MN 615 ¼, 125 mm) were purchased from Macherey-Nagel. Syringe filters (0.45 μm, PA) were purchased from Carl Roth. (−)-Quinic acid was purchased from Thermo Fisher Kandel (Karlsruhe, Germany).
Plant Material and Insect Larvae
Plant samples of black poplar (P. nigra) were collected from trees growing in proximity to the Max Planck Institute for Chemical Ecology in Jena, Germany. P. nigra is a typical hostplant of C. vinula (Hintze-Podufal 1970) in natural habitats and was therefore used to raise the larvae for the experiments. P. trichocarpa x deltoides Beaupré were grown in the greenhouse of the Max Planck Institute for Chemical Ecology. The species was thoroughly examined in previous studies regarding its spectrum of salicinoid defense compounds (Feistel et al. 2015). The light period was set from 6:30 to 20:30 (14 h), while temperatures were kept between 21 and 23 °C during the day and between 19 and 21 °C at night. The humidity was regulated between 50 to 60%. Puss moth (C. vinula) larvae were hatched from eggs and reared on P. nigra leaves in the laboratory.
Extraction and Isolation of C. vinula Frass.
Frass of C. vinula larvae fed on P. nigra leaves were collected and lyophilized, resulting in 73 g of dry material. Contaminated material (leaves, petioles and exuviae) was removed manually. Dried frass (20 g) were ground using a ceramic mortar and pestle, and extracted (5 × 200 ml, each 10 min) with MeOH. The extracts were filtered (paper filters and 0.45 μm PA syringe filters), pooled and evaporated under reduced pressure, resulting in 2.5 g dried crude extract. This dry matter (53.8 mg) was suspended in water (20 ml) using ultrasound and subjected to pre-separation on a HR-X SPE (PS/DVB) cartridge. After conditioning with MeOH (2 × 6 ml) and equilibration with water (3 × 6 ml), the cartridge was loaded with the extract suspension and washed with water (3 × 6 ml). After drying in vacuum, the cartridge was eluted with MeOH (3 × 6 ml). The eluate was then dried using a vacuum centrifuge, resulting in 24.7 mg pre-purified extract. For separation, an aliquot dissolved in MeOH (67.4 mg ml−1) was subjected to HPLC-SPE. A binary solvent system of 0.1% formic acid in water (solvent A) and 0.1% formic acid in MeOH (solvent B) was used for HPLC separation, starting with a 5 min isocratic flow of 100% solvent A and decreasing linearly for 90 min to 50% solvent A. Column temperature was set to 35 °C and the solvent flow rate was 0.8 mL min−1. After each run, the column was washed with 100% MeOH for 5 min and equilibrated with 100% H2O for 10 min. SPE cartridges loaded with metabolites were dried in a stream of N2 gas before being eluted with MeOH. Eluted compounds were dried by vacuum centrifugation, yielding the following compounds (Rt retention time): 1 (Rt 32.69 min, 2.1 mg g−1 dry frass), 4 (Rt 36.69 min, 1.2 mg g−1), 5 (Rt 39.71 min, 1.1 mg g−1), 2 (Rt 41.00 min, 2.6 mg g−1), 3 (Rt 52.59 min, 3.4 mg g−1), 10 (Rt 61.70 min, 1.6 mg g−1), 7 (Rt 64.14 min, 2.1 mg g−1), 9 (Rt 65.52 min, 3.0 mg g−1), 6 (Rt 68.34 min, 2.8 mg g−1), 11 (Rt 70.18 min, 2.1 mg g−1), 8 (Rt 72.73 min, 1.9 mg g−1).
Structure elucidation was carried out using 1D (1H NMR, selective TOCSY) and 2D NMR (1H-1H COSY, 1H-13C HSQC, 1H-13C HMBC) spectra recorded at 700 MHz and UHPLC-ESI-MS/MS measurements (Q Exactive Plus - Orbitrap MS) using a binary solvent system of H2O (solvent A) and acetonitrile (solvent B), both of which contained 0.1% (v/v) formic acid with a flow rate of 300 μl min−1. The linear gradient used started with 0% B and increased to 100% B within 15 min. Afterwards, the column was washed for 5 min with 100% B and then equilibrated at 0% B for 5 min.
In Vivo Generation and Isolation of 13C-Labeled Salicortin
Stable isotope labeling was achieved in a growth chamber resembling a setup described previously (Chen et al. 2011). The greenhouse light system (Philips SON-T Agro 400 W) was used to provide constant light exposure from 6:30 to 22:00 (15.5 h). Temperature and relative humidity were kept between 20 °C to 30 °C and 50% to 80%, respectively. Six P. trichocarpa x deltoides Beaupré plants were pruned to a height of about 30 cm, leaving a few basal leaves. After being transferred to the growth chamber, plants were kept in darkness for the first 2 days. At the beginning of day 3, the respired CO2 (natural abundance isotope ratio) was removed from the chamber’s atmosphere and 450 ppm 13CO2 was injected into it. This 13CO2 level was kept constant during the entire experimental time (26 days). At the end of each day’s light period, 13CO2 injection was stopped and respired CO2 was continuously removed during the night. Details about the 13CO2 labeling are provided in the supporting information (B.1 and B.2).
The 13CO2-labeling experiment was stopped at day 28, and newly grown plant tissue (leaves with petioles) was collected and lyophilized yielding 13 g dry material. The material was manually crushed and filled equally into Falcon tubes (8 × 45 ml) containing steel beads (3 mm). Extraction was accomplished with MeOH (30 ml per tube) in a Skandex S-7 paint shaker (Fluid Management, Wheeling, IL, USA) for 2 × 4 min. Afterwards, the extracts were pooled, filtered, and the solvent was evaporated in vacuo, yielding 4.18 g (32% dw) dry matter. The dry matter was suspended in 500 ml H2O, divided into 3 parts and subjected to solid-phase extraction on HR-X SPE columns (each 1.5 g sorbent). After loading, columns were washed with H2O (3 × 20 ml) and eluted with MeOH (20 ml). The combined methanolic fractions were dried in vacuo, yielding 2.26 g pre-purified extract (17.33% dw). An aliquot solution (121 mg ml−1) was subjected to HPLC separation.
[U-13C]Salicortin was separated using a binary solvent system consisting of 0.1% formic acid in H2O (solvent A) and 0.1% formic acid in MeOH (solvent B). Column temperature was set to 35 °C and the solvent flow rate was 0.8 ml min−1. The HPLC gradient that was used started with a 5 min isocratic flow of 100% solvent A and decreased linearly for 5 min to 85%, 25 min to 70% and finally 50 min to 50% solvent A. Afterwards, the column was washed for 10 min with 100% MeOH and equilibrated for 10 min with 100% H2O. The salicortin UV signal peak (λ = 285 nm) appeared at Rt 42.6 min. Unlabeled salicortin (natural abundance 13C) was isolated in the same manner from unlabeled P. trichocarpa x deltoides tissue.
Characterization of the salicortin isolated from the 13C-labeled plant tissue and calculation of its 13C-enrichment was done by means of 1H, 13C and 1H-13C HSQC NMR spectra (400 MHz) and HPLC-ESI-MS measurements using the chromatographic method as described above. The in vivo-generated salicortin showed uniform 13C-labeling with 82% total 13C-enrichment. Spectroscopic data of the [U-13C]salicortin (82% 13C) and detailed information about the calculation of the 13C-enrichment are presented in the Supplementary data (SI B.2).
[U-13C]Salicortin Larval Feeding
Freshly cut leaves of P. nigra (LPI 3–10) were used for feeding experiments. The leaf petioles were inserted into 2 ml Eppendorf micro reaction vessels. Lids of the vessels were removed for convenient handling prior to being filled with tap water. The opening of the vessels was sealed with Parafilm® fastening the leaf petioles to prevent the water in the vessel from spilling. An aqueous solution of [U-13C]salicortin (2.5 mg ml−1) was spotted evenly on the surface of 5 poplar leaves (10 × 20 μl droplets per leaf). Control leaves were spotted with water in an analogous fashion. Leaves were left under the fume hood for 3 h, allowing the droplets to dry completely. Subsequently, the [U-13C]salicortin-coated leaves and the control leaves were transferred into arenas for feeding experiments. Each arena contained one leaf and one C. vinula larva (late 3rd to early 4th instars), which were placed onto the surface of each leaf. The larvae were kept in the arena until the leaves had been completely consumed. Frass collected from the control experiments and the stable isotope feeding experiments, respectively, were pooled into 2 batches which were subsequently lyophilized.
The freeze-dried frass was then extracted with MeOH (3 × 1 ml) by homogenization in a Minilys cell disruptor (2 ml tubes, 60 s and 4000 rpm), using 1.4 mm o.d. ZrO2 beads. The supernatants from each sample were pooled and evaporated using N2 gas and subsequently extracted with H2O (5 × 1 ml). The aqueous extracts were pooled and dried with N2 gas. Afterwards, methanolic and aqueous extracts of frass from control and [U-13C]salicortin feeding experiments were subjected to NMR (500 MHz) and UHPLC-ESI-MS/MS (Q Exactive Plus – Orbitrap MS) analysis, using the chromatographic system described above.
Collection of C. vinula Hemolymph
The question of whether salicinoid metabolites are detectable in the hemolymph of C. vinula larvae was addressed as follows: 6 C. vinula larvae (3 × control, 3 × fed with [U-13C]salicortin) were immobilized in 15 ml Falcon tubes and kept in the −20 °C freezer for 15 min. A mid-abdominal proleg of an anesthetized larva was pierced with a scalpel, and the emerging hemolymph was transferred by means of a pipette into an ice-cooled 2 ml Eppendorf micro reaction vessel containing MeOH (1 ml). Subsequently, the mixture was centrifuged and the supernatant was evaporated by a stream of N2 gas, yielding an average amount of 3.07 mg (SD ± 1.51 mg) residue per larva. Each of the 6 dried residues were dissolved in 1 ml MeOH and analyzed independently by UHPLC-ESI-MS/MS (LTQ - Orbitrap XL MS), using a binary solvent system of H2O (solvent A) and acetonitrile (solvent B), both of which contained 0.1% (v/v) formic acid with a flow rate of 300 μl min−1. A linear gradient was used, starting with 5% B to 52% B within 20 min. Afterwards, the column was washed for 5 min with 100% B and then equilibrated at 5% B for another 5 min.
Results
C. vinula Frass Analysis
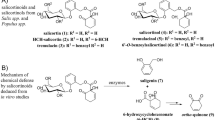
Frass of C. vinula larvae fed on P. nigra leaves was collected and metabolites extracted as described in the Methods and Materials section. Eleven quinic acid derivatives representing almost 2.5% (23.9 mg g−1) of the frass dry mass were identified (Fig. 1). Among them, 10 compounds (1–3, 5–11) were unknown and only compound 4 had been reported recently (Wan et al. 2016). For an overview about NMR, HR-MS and UV data, see Tables 1, 2, 3 and 4. Spectra of all compounds, including salicortin (12) and quinic acid (13), are provided in the Supplementary data (SI A.1 to A.13).
The HRESIMS data of compound 1 showed a molecular ion peak at m/z 311.0774 [M-H]− corresponding to a molecular formula of C14H16O8 (calcd for C14H15O8, m/z 311.0772). The 1H NMR spectrum of compound 1 (Table 1) showed signals of 11 protons assignable to 2 different structural units. In the low-field region of the 1H NMR spectra, we observed signals of an asymmetric 4-spin system (ABCD) at δH 6.62 (3JHH = 0.8/8.3 Hz; H-3′), δH 7.21 (3JHH = 1.5/7.3/8.3 Hz; H-4′), δH 6.60 (3JHH = 0.8/6.9/7.3 Hz; H-5′) and δH 7.44 (3JHH = 1.4/6.9 Hz; H-6′), characteristic of a 1,2-disubstituted aromatic ring. The corresponding 13C chemical shifts were determined by an 1H-13C hetero-correlation single quantum coherence (HSQC) spectrum as δC 118.8 (C-3′), δC 135.5 (C-4′), δC 119.5 (C-5′) and δC 132.5 (C-6′). Further evidence for a 1,2-disubstitution was provided by an 1H-13C hetero-nuclear multiple-bond correlation (HMBC) spectrum showing 3JCH correlations from H-4′ and H-6′ to a quaternary carbon atom at δC 159.9 (C-2′) as well as from H-3′ and H-5′ to another quaternary carbon atom at δC 115.2 (C-1′). Because of its low-field 13C chemical shift, C-2′ was assigned as oxygenated. Furthermore, H-6′ (δH 7.44) showed a long-range CH-correlation to a carboxyl functionality at δC 167.7 (C-7′) tethered to C-1′ (δC 115.2). Thus, the data suggest the presence of a salicyloyl moiety (SA). A double-doublet signal of a methine appeared at δH 5.70 (3JHH = 3.1/3.4/3.1; H-3). The corresponding 13C chemical shift was extracted from the 1H-13C HSQC spectrum at δC 72.8 (C-3), which is characteristic for a hydroxylated aliphatic carbon atom. Adjacent to H-3, another methine at δH 3.60 (3JHH = 3.4/9.5 Hz; H-4), and a methylene group at δH 2.35 (3JHH = 3.1/14.4 Hz; H-2a) and δH 2.11 (3JHH = 3.1/14.4 Hz; H-2b) were determined from their 3JHH correlations by 1H-1H COSY. Furthermore, consecutive cross-peaks from H-4 to another methine at δH 4.06 (3JHH = 4.6/9.5/12.0 Hz; H-5) and to a second methylene group at δH 2.10 (3JHH = 12.0/12.5 Hz; H-6a) and δH 1.95 (3JHH = 4.6/12.5 Hz; H-2b) were observed. The small 3JHH values (3.4 Hz) for H-3 and H-4 indicated equatorial configuration, whereas the large values (9.5 Hz) for H-4 and H-5 indicated axial configuration. The corresponding 13C-chemical shifts were determined by 1H-13C HSQC as δC 36.8 (C-2), δC 75.6 (C-4), δC 68.0 (C-5) and δC 42.5 (C-6). As for C-3 (δC 72.8), large 13C NMR chemical shift values indicated oxygenation for C-4 and C-5. The1H-13C HMBC spectrum revealed a 3JCH correlation of H-3 with an oxygenated quaternary carbon atom at δC 80.3 (C-1). Furthermore, both methylene groups (H-2ab and H-6ab) showed weak long-range CH correlations to C-1, as a result of which the entire aliphatic structure can be characterized as a cyclohexane ring system. Another HMBC correlation from H-6b to a quaternary carbon at δC 182.6 (C-7) tethered to C-1 revealed quinic acid. The characteristic low-field shift of H-3 indicated substitution in this position. This assumption was further supported by an 1H-13C HMBC correlation from H-3 to the carboxyl carbon C-7′ (δC 167.7) of the salicyloyl moiety. Accordingly, the structure of compound 1 was assigned as 3-O-salicyloyl quinic acid.
Similar to 1, the HRESIMS spectra of compounds 2 and 3 showed a molecular ion peak of m/z 311.0773 [M-H]−, again corresponding to a molecular formula of C14H16O8 (calcd for C14H15O8, m/z 311.0772). The data from 1H NMR, 1H-1H COSY, 1H-13C HSQC and the 1H-13C HMBC spectra resembled those of compound 1 (Table 1). Signals of the salicyloyl moiety and the quinic acid subunit were present, but the substitution patterns differed at C-3, C-4 and C-5 of the quinic acid moieties. Acylation of the hydroxyl groups in (−)-quinic acid (13) leads to low-field shifts of the signals of H-3, H-4 and H-5 (Pauli et al. 1998). However, the multiplicities and coupling constants remain unaffected. Thus, the shift of a characteristic multiplet to the low field indicates the substitution site (Fig. 2). The multiplets of H-4 (δH 5.02) in the 1H NMR spectrum of compound 2 and H-5 (δH 5.55) in the spectrum of compound 3 – which in comparison with the corresponding signals of quinic acid (13) appeared at the low field – revealed esterification with salicyloyl units at the hydroxyl groups in positions 4 and 5, respectively.
The structures of 2 and 3 were further characterized by 1H-13C HMBC correlations of H-4 (2; δH 5.02) and H-5 (3; δH 5.55) with the carboxylic carbon atoms of their salicyloyl moieties at δC 170.8 (2; C-7′) and 170.7 (3; C-7′), respectively. Accordingly, compound 2 was identified as 4-O-salicyloyl quinic acid and compound 3 was identified as 5-O-salicyloyl quinic acid.
The analytical data of compound 4 were in accordance with previously reported 4-O-benzoyl quinic acid (Wan et al. 2016). The HRESIMS spectra of compound 5 showed a molecular ion peak of m/z 295.0819 [M-H]−, corresponding to a molecular formula of C14H16O7 (calcd for C14H15O7, m/z 295.0823) and indicating an isomer of compound 4. In accordance with this suggestion, the 1H NMR, 1H-1H COSY, 1H-13C HSQC and the 1H-13C HMBC spectra of compound 5 (Table 2) showed very similar signals in comparison to compound 4. Thus, both compounds are constitutional isomers consisting of a quinic acid moiety substituted with a benzoyl moiety. Accordingly, the 1H NMR data of compound 5 showed signals assignable to a symmetrical 5-spin system (AA’XX’Y) at δH 8.06 (3JHH = 1.2/8.3 Hz, H-2′/6′), δH 7.47 (3JHH = 7.4/8.3 Hz, H-3′/5′) and δH 7.59 (3JHH = 7.4/7.4 Hz, H-4′); these signals are characteristic of a mono-substituted aromatic ring. Corresponding 13C chemicals shifts were extracted from the 1H-13C HSQC spectrum at δC 130.5 (C-2′/6′), δC 129.4 (C-3′/5′) and δC 134.1 (C-4′). The aromatic ring structure was further determined by 3JCH correlations in the 1H-13C HMBC spectrum from H-3′/5′ (δH 7.47) to a quaternary carbon atom at δC 131.6 (C-1′). Another long-range CH-correlation of H-2′/6′ (δH 8.06) to a carboxyl carbon atom at δC 168.1 (C-7′) tethered to the C-1′ position of the aromatic ring was observed in the HMBC spectrum, confirming the benzoyl moiety. Substitution to the quinic acid moiety was determined by the characteristic low-field shift of the signal of H-5 (δH 5.52), indicating substitution at the hydroxyl group at position 5 (δC 73.3; δH 5.52). Accordingly, the structure of compound 5 was assigned as 5-O-benzoyl quinic acid.
The HRESIMS data of compound 6 showed a molecular ion peak of m/z 431.0986 [M-H]−, corresponding to a molecular formula of C21H20O10 (calcd for C21H19O10, m/z 431.0984). The 1H NMR, 1H-1H COSY, 1H-13C HSQC and the 1H-13C HMBC spectra of compound 6 (Table 2) showed signals of a quinic acid as well as 2 salicyloyl moieties. The 1H NMR spectra showed characteristic low-field shifts for H-3 (δH 5.95) and H-4 (δH 5.28), suggesting a bis-substituted quinate. Due to overlapping signals, selective TOCSY together with 1H-1H COSY and 1H-13C HSQC were employed to extract the chemical shifts of both salicyloyl moieties. Connectivities between quinate and salicyloyl moieties were established by 1H-13C long-range correlations of H-3 to δC 169.3 (C-7′) and of H-4 to δC 170.6 (C-7″), both characterized as carboxyl carbons of the respective salicyloyl units. Accordingly, the structure of compound 6 was assigned as 3-O,4-O-disalicyloyl quinic acid.
The HRESIMS spectra of compounds 7 and 8 showed molecular ion peaks of m/z 431.0985 [M-H]− and 431.0987 [M-H]−, respectively. As in compound 6, both values corresponded to a molecular formula of C21H20O10 (calcd for C21H19O10, m/z 431.0984). 1H NMR, 1H-1H COSY, 1H-13C HSQC and the 1H-13C HMBC spectra showed almost the same signals (Table 2), indicating quinic acid esters with salicyloyl moieties as substituents. The structures were characterized by 1H-13C long-range correlations as well as the characteristic chemical shifts of 1H NMR signals for H-3, H-4 and H-5. For compound 7, we observed low-field shifts for H-3 (δH 5.708) and H-5 (δH 5.712), suggesting acylation at this positions. The assumption was proven by 3JCH correlations of H-3 to δC 169.8 (C-7′) and of H-5 to δC 170.3 (C-7″), indicating C-7′ and C-7″ as carboxyl carbons of 2 different salicyloyl moieties. Likewise, for compound 8, we observed low-field shifts for H-4 at δH 5.42, which further showed a 3JCH correlation to δC 170.5 (C-7′). The 1H NMR signal of H-5 appeared at δH 5.95 with a 3JCH correlation to δC 170.4 (C-7″). Accordingly, the structures of the compounds 7 and 8 were assigned as 3-O,5-O-disalicyloyl quinic acid and 4-O,5-O-disalicyloyl quinic acid, respectively.
The HRESIMS spectra of compound 9 showed a molecular ion peak of m/z 415.1038 [M-H]− corresponding to a molecular formula of C21H20O9 (calcd for C21H19O9, m/z 415.1035). The NMR data (Table 3) extracted from the 1H NMR, 1H-1H COSY, 1H-13C HSQC and the 1H-13C HMBC spectra were very similar to those of compounds 1 and 6. Accordingly, the presence of a salicyloyl quinate unit was suggested. Furthermore, signals of a benzoyl moiety as described for compound 4 were observed as a substituent of quinic acid. The 1H NMR spectra showed characteristic low-field shifts for H-3 (δH 5.93) and H-4 (δH 5.22), suggesting substitution at those positions. Cross-signals observed in the 1H-13C HMBC between H-3 and the carboxylic carbon atom C-7′ (δC 169.5) of the salicyloyl moiety proved the substitution in position C-3 of the quinic acid via an ester bond. An additional 3JCH correlation of H-4 to the carboxylic carbon atom C-7″ (δC 167.2) of the benzoyl moiety revealed another esterification. Accordingly, the structure of compound 9 was assigned as 3-O-salicyloyl-4-O-benzoyl quinic acid.
Like the data for compound 9, the HRESIMS data for compounds 10 and 11 showed a molecular ion peak of 415.1038 [M-H]− corresponding to a molecular formula of C21H20O9 (calcd for C21H19O9, m/z 415.1035). The NMR data for both compounds (Table 3) were very similar to those of 9, suggesting acylated quinic acid derivatives with mixed salicyloyl and benzoyl substituents. The structures of compounds 10 and 11 were elucidated by characteristic 1H NMR chemical shifts and 1H-13C long-range correlations as described for compounds 6–8. For compound 10, substitution with a salicyloyl group in position 3 (δC 74.4; δH 5.71) and with the benzoyl unit in position 5 (δC 72.9; δH 5.56) of the quinic acid was observed. For compound 11, salicyloyl substitution was observed at position 4 (δC 77.6; δH 5.41), and benzoylation was found at position 5 (δC 69.8; δH 5.91) of the quinic acid moiety. Accordingly, the structures were assigned as 3-O-salicyloyl-5-O-benzoyl quinic acid (10) and 4-O-salicyloyl-5-O-benzoyl quinic acid (11).
Hemolymph Analysis
Hemolymph samples of 6 C. vinula larvae were analyzed by UHPLC-ESI-MS/MS. Mass spectra were scanned for the presence of the molecular ions of compounds 1–11 (m/z 311, 295, 431, 415 ± 0.5 [M-H]−) as well as for the molecular ions of salicortin (12, m/z 424 ± 0.5 [M-H]−) and salicin (m/z 285 ± 0.5 [M-H]−). None of these compounds were detected. Data are provided in Supplementary data (SI D.1 and D.2).
13C-Labeling Study
In order to investigate a possible precursor-product relationship of salicortin with the metabolites 1–11, leaves of P. nigra were spotted with [U-13C]salicortin (82% 13C), as described in the Methods and Materials section (2.5), and subsequently fed to C. vinula larvae. The frass of larvae were extracted and analyzed by NMR and UHPLC-HRMS (see Supplementary data, C.2 and C.3). All larvae survived the feeding experiment without exhibiting negative effects arising from an elevated salicortin intake.
The 13C NMR spectrum of the frass extract from the [U-13C]salicortin feeding experiment showed signals characteristic for compounds 1–11. Pronounced 13C satellites appeared for signals of salicylates around δC 170, δC 165 and δC 135–130. No 13C enrichment was found for glucosyl or quinate moieties (SI C.2–1 to C.2–5). Further structure elucidation was accomplished by 2D 1H-13C correlation spectroscopy. 1H-13C HSQC allowed for the identification of protons attached to the 13C-enriched positions. The associated spin systems were assigned by selective TOCSY experiments to δH 7.93 (dd, 3JHH = 1.8/8.0 Hz)/δC 131.0, δH 7.98 (dd, 3JHH = 1.9/8.0 Hz)/δC 131.7 and δH 8.04 (3JHH = 1.8/8.0 Hz)/δC 131.3, corresponding to salicylic acid moieties such as those observed for compounds 1–3 and 6–11. The 1H-13C HMBC spectrum showed correlations of the salicylic acid moiety protons at δH 7.93 (δC 131.0; C-6), δH 7.98 (δC 131.6; C-6) and δH 8.04 (δC 131.3; C-6), with carbon atoms resonating at δC 136.7/136.5/136.2 (C-4), δC 162.1/162.8 (C-2) and δC 171.0/171.2/171.5 (C-7; COOH). 13C-13C Satellite signals for all those carbon signals indicated multiple 13C enrichment of the salicyloyl moieties.
No 13C-13C satellites were observed in the 13C NMR spectra and thus no 13C enrichment occurred in the benzoyl units. The low intensity of the 1H-13C HMBC correlations of the benzoyl protons at δH 8.09 (δC 130.5; C-2/6) and δH 8.13 (δC 130.6; C-2/6) with δC 134.0 (C-4) and δC 168.1 (C-7; COOH) (Fig. 3 and SI C.2–6 to C.2–17) confirmed that the benzoyl units remained unlabeled.
Superimposed 1H-13C HSQC (black) and HMBC spectra (red) (500 MHz, MeOH-d4) of C. vinula frass extract demonstrating 13C enrichment of salicyloyl (SA) units but not of benzoyl (BA) units from [U-13C]salicortin feeding. Top: Partial 1H NMR spectrum showing signals of BA and SA. In the 13C NMR spectrum (125 MHz) on the left, resonances of salicyloyl substituents show coupling patterns indicative of 13C-enrichment. Cross-signals of * are not shown
Furthermore, UHPLC-HRMS spectra of the frass extract of C. vinula larvae fed with [U-13C]salicortin showed 13C-isotopologue patterns from [M-H + 3]− up to [M-H + 7]− (Fig. 4; C.3–1 to C.3–23). Remarkably, only salicylate-containing quinate esters, mono-salicyloyl quinates 1–3, di-salicyloyl quinates 6–8 and salicyloyl-benzoyl quinates 9–11, were labeled, and benzoyl quinates 4 and 5 showed no 13C enrichment. The incorporation of 13C from [U-13C]salicortin into the salicyloyl moiety of the acylated quinic acids 1–3 and 6–11 clearly indicated a substrate-product relationship. Accordingly, we concluded that the acylated quinic acids were downstream products of the salicortin metabolism in C. vinula.
Superimposed extracted ion HRMS spectra of the 4 acylated quinic acid groups – mono-salicyloyl quinates, mono-benzoyl quinates, di-salicyloyl quinates and salicyloyl-benzoyl quinates – isolated from the frass of C. vinula larva reared on [U-13C]salicortin-coated P. nigra leaves (red) and unlabeled leaves (black). The groups are represented by compounds 1 (A), 4 (B), 6 (C) and 9 (D). The gray labeled isotopologue signals in panel C are very weak and thus not visible in this magnification
Another 7 (up to [M-H + 14]−) isotopologue peaks are present in the spectra of di-salicyloyl quinates 6–8. The [M-H + 8]− peak may be due to the presence of a natural abundance 13C1 quinate or a 13C1 salicyloyl substituent, in addition to the 13C7-labeled first salicyloyl unit.
Discussion
The chemical defense of poplar includes abundant phenolic glycosides, such as salicortin and tremulacin, which can make up to 4% of the leaf dry mass (Boeckler et al. 2011; Donaldson et al. 2006). It is widely accepted that the digestive activation of salicinoids leads to toxic products warding off non-adapted herbivores. For salicortin, it has been proposed that activation proceeds through deglucosylation by β-glucosidases (Clausen et al. 1990; Lindroth 1988; Pentzold et al. 2014). The aglycon is either hydrolyzed spontaneously in the alkaline gut environment or degraded enzymatically by insect esterases (Lindroth et al. 1988; Lindroth 1989) to saligenin and a 1-hydroxy-6-oxocyclohex-2-en-1-oyl (HCH) fragment (Haruta et al. 2001; Julkunen-Tiitto and Meier 1992). The latter is believed to be oxidized to pyrocatechol and finally to o-quinone (Appel 1993; Barbehenn et al. 2010; Knuth et al. 2011; Ruuhola et al. 2003). o-Quinones have a high potential to bind to a variety of biomolecules, including amino acids and proteins (Haruta et al. 2001; Smith 1985). Generalist lepidopteran species, such as the gypsy moth (L. dispar), have developed a strategy of detoxification (Boeckler et al. 2016) where the major salicinoids in P. nigra, salicortin and tremulacin, are transformed into typical metabolic phase II conjugates (Grant 1991), with salicin as the major metabolite (Boeckler et al. 2016).
The compounds 1–11 described in the present work are new quinic acid derivatives, isolated from the frass of C. vinula. The quinic acid moiety is substituted with either 1 or 2 salicylate units (1–3, 6–8), benzoate units (4, 5) or one of each unit (9–11). Compound 4 is the only compound that has been previously reported, namely as a constituent of fruits of Ficus hirta (Wan 2016). The results show that conjugation with quinic acid plays a decisive role in the transformation of salicinoids by C. vinula. The conjugation with quinic acids is described here for the first time as part of a detoxification pathway. Quinic acid has been reported to play a role in response to herbivory (Wang et al. 2016), and quinic acids were detected in the gut of insects (Crecelius et al. 2017). It was also suggested that quinic acid conjugates like chlorogenic acid produce feeding-deterrent compounds after cleavage and oxidative transformation of the phenylpropanoic part (Stevenson et al. 1993). The latter aspect needs to be clarified in the present example, and experiments addressing the question are ongoing.
Unlike in L. dispar, deglycosylation is an important feature of the metabolism in C. vinula, as glycosidic compounds or free sugars are absent in the frass extract. Generally, the salicinoid breakdown follows reported routes like deglucosylation, ester cleavage and conjugation (Clausen et al. 1990; Lindroth 1988; Pentzold et al. 2014). The structural diversity of the compounds – namely, the differential substitution of quinic acid with salicylate or benzoate – can be rationalized by acyl migration under basic conditions as present in the insect gut (Appel and Martin 1990). An enzymatic and hence stereospecific conjugation with quinic acid may take place at first, but those products rapidly isomerize later on. Another metabolite present after salicinoid degradation in L. dispar, hippuric acid, was absent in the [13C]-metabolite spectrum of C. vinula. We can therefore conclude that hippuric acid is not a degradation product of salicortin, but arises likely from benzoyl-substituted structures like tremulacin.
The question of whether the source of salicyloyl units in quinic acid derivatives 1–11 were indeed salicinoid glycosides was tackled by stable isotope labeling experiments. C. vinula larvae were allowed to feed on leaves of P. nigra spotted with [U-13C]salicortin (82% 13C). Frass extracts were analyzed by means of NMR and HRMS. The resulting data were screened for isotopic incorporation into 1–11. The isotopologue pattern of both NMR and HRMS spectra indicated differences in the incorporation of labeled salicortin into mono-salicyloyl quinic acids (1–3), mono-benzoyl quinic acids (4, 5), bis-salicyloyl quinic acids (6–8) and salicyloyl-benzoyl quinic acids (9–11). NMR data showed enrichment of 13C only in the salicyloyl moiety of quinic acid derivatives (Fig. 3). 13C enrichment was further supported by HRMS data (Fig. 4) which showed a specific isotopologue pattern spanning from [M-H]− to [M-H + 7]− for 1–3. The HRMS spectra of 4 and 5 did not show additional 13C-isotopologue signals. Thus, neither quinic acid nor benzoyl moieties is thought to represent a downstream product of the [U-13C]salicortin precursor. This suggestion was further reflected in the HRMS data of 6–11. Whereas mixed salicyloyl-benzoyl substituted compounds show only isotopologue signals up to [M-H + 7]−, the bis-salicyloyl derivatives show signals up to [M-H + 14]−.
Accordingly, this finding is a clear evidence that saligenin is transformed into salicylic acid. We assume that structurally similar salicinoids are transformed in an analogous manner. No 13C incorporation was detected in the quinic acid moieties of 1–11. Therefore, quinic acids likely originate from chlorogenic acids (caffeoyl quinates), which have already been described as leaf constituents of the Salicaceae (Caseys et al. 2015; Glynn et al. 2004). Considering the metabolic scheme in Fig. 5, it is important to note that there seems to be only one specific oxidative transformation of saligenin into salicylic acid in C. vinula. Likely, this specificity prevents the caffeoyl moiety of chlorogenic acid to be transformed into a toxic species (Felton et al. 1989) and facilitates quinic acids to scavenge the phenolic degradation products.
In our search for break-down products related to the HCH-moiety of salicortin, we did not find any further 13C-labeled compound. Therefore, we speculate that there is a mechanism by which HCH degradation yields salicylic acid (Fig. 5). Under basic conditions, the HCH fragment could rearrange to an HCH-enolate followed by a spontaneous dehydration to form salicylic acid. The feasibility of such a transformation was observed in synthetic studies (Nagasawa et al. 2010). The origin of benzoyl substituents in 4, 5, 9, 10 and 11 are likely common Salicaceae leaf constituents, such as chaenomeloidin, nigracin, populin, salireposid, tremulacin or tremuloidin (Boeckler et al. 2011).
References
Ali JG, Agrawal AA (2012) Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci 17:293–302
Appel HM (1993) Phenolics in ecological interactions: the importance of oxidation. J Chem Ecol 19:1521–1552
Appel HM, Martin MM (1990) Gut redox conditions in herbivorous lepidopteran larvae. J Chem Ecol 16:3277–3290
Barbehenn R, Dukatz C, Holt C, Reese A, Martiskainen O, Salminen J-P, Yip L, Tran L, Constabel CP (2010) Feeding on poplar leaves by caterpillars potentiates foliar peroxidase action in their guts and increases plant resistance. Oecologia 164:993–1004
Beran F, Pauchet Y, Kunert G, Reichelt M, Wielsch N, Vogel H, Reinecke A, Svatoš A, Mewis I, Schmid D, Ramasamy S, Ulrichs C, Hansson BS, Gershenzon J, Heckel DG (2014) Phyllotreta striolata flea beetles utilize host plant defense compounds to create their own glucosinolate-myrosinase system. Proc Natl Acad Sci U S A 111:7349–7354
Boeckler GA, Gershenzon J, Unsicker SB (2011) Phenolic glycosides of the salicaceae and their role as anti-herbivore defenses. Phytochemistry 72:1497–1509
Boeckler GA, Paetz C, Feibicke P, Gershenzon J, Unsicker SB (2016) Metabolism of poplar salicinoids by the generalist herbivore Lymantria dispar (Lepidoptera). Insect Biochem Mol Biol 78:39–49
Burse A, Frick S, Discher S, Tolzin-Banasch K, Kirsch R, Strauß A, Kunert M, Boland W (2009) Always being well prepared for defense: the production of deterrents by juvenile Chrysomelina beetles (Chrysomelidae). Phytochemistry 70:1899–1909
Caseys C, Stritt C, Glauser G, Blanchard T, Lexer C (2015) Effects of hybridization and evolutionary constraints on secondary metabolites: the genetic architecture of phenylpropanoids in European Populus species. PLoS One 10:23
Chen W-P, Yang X-Y, Harms GL, Gray WM, Hegeman AD, Cohen JD (2011) An automated growth enclosure for metabolic labeling of Arabidopsis thaliana with 13C-carbon dioxide - an in vivo labeling system for proteomics and metabolomics research. Proteome Sci 9:9
Clausen TP, Reichardt PB, Bryant JP, Werner RA, Post K, Frisby K (1989) Chemical model for short-term induction in quaking aspen (Populus tremuloides) foliage against herbivores. J Chem Ecol 15:2335–2346
Clausen TP, Koller JW, Reichardt PB (1990) Aglycone fragmentation accompanies β-glucosidase catalyzed hydrolysis of salicortin, a naturally-occurring phenol glycoside. Tetrahedron Lett 31:4537–4538
Crecelius AC, Michalzik B, Potthast K, Meyer S, Schubert US (2017) Tracing the fate and transport of secondary plant metabolites in a laboratory mesocosm experiment by employing mass spectrometric imaging. Anal Bioanal Chem 409:3807–3820
Donaldson JR, Stevens MT, Barnhill HR, Lindroth RL (2006) Age-related shifts in leaf chemistry of clonal aspen (Populus tremuloides). J Chem Ecol 32:1415–1429
Feistel F, Paetz C, Lorenz S, Schneider B (2015) The absolute configuration of salicortin, HCH-salicortin and tremulacin from Populus trichocarpa x deltoides Beaupré. Molecules 20:5566–5573
Felton GW, Donato K, Del Vecchio RJ, Duffey SS (1989) Activation of plant foliar oxidases by insect feeding reduces nutritive quality of foliage for noctuid herbivores. J Chem Ecol 15:2667–2694
Glynn C, Ronnberg-Wastljung AC, Julkunen-Tiitto R, Weih M (2004) Willow genotype, but not drought treatment, affects foliar phenolic concentrations and leaf-beetle resistance. Entomol Exp Appl 113:1–14
Grant DM (1991) Detoxification pathways in the liver. J Inherited Metab Dis 14:421–430
Haruta M, Pedersen JA, Constabel CP (2001) Polyphenol oxidase and herbivore defense in trembling aspen (Populus tremuloides): cDNA cloning, expression, and potential substrates. Physiol Plant 112:552–558
Hintze-Podufal C (1970) Über die quantitativen Änderungen der Kotabgabe während der Larvalentwicklung von Cerura vinula L. (Lepidoptera). Oecologia 5:334–346
Joussen N, Agnolet S, Lorenz S, Schone SE, Ellinger R, Schneider B, Heckel DG (2012) Resistance of Australian Helicoverpa armigera to fenvalerate is due to the chimeric P450 enzyme CYP337B3. Proc Natl Acad Sci USA 109:15206–15211
Julkunen-Tiitto R, Meier B (1992) The enzymatic decomposition of salicin and its derivatives obtained from salicaceae species. J Nat Prod 55(9):1204–1212
Knuth S, Schübel H, Hellemann M, Jürgenliemk G (2011) Catechol, a bioactive degradation product of salicortin, reduces TNF-α induced ICAM-1 expression in human endothelial cells. Planta Med 77:1024–1026
Lindroth RL (1988) Hydrolysis of phenolic glycosides by midgut β-glucosidases in Papilio glaucus subspecies. Insect Biochem 18:789–792
Lindroth RL (1989) Biochemical detoxication: mechanism of differential tiger swallowtail tolerance to phenolic glycosides. Oecologia 81:219–224
Lindroth RL (1991) Biochemical ecology of aspen-Lepidoptera interactions. J Kans Entomol Soc 64:372–380
Lindroth RL, Hemming JDC (1990) Responses of the gypsy moth (Lepidoptera: Lymantriidae) to tremulacin, an aspen phenolic glycoside. Environ Entomol 19:842–847
Lindroth RL, Scriber JM, Hsia MTS (1988) Chemical ecology of the Tiger swallowtail - mediation of host use by phenolic glycosides. Ecology 69:814–822
Nagasawa T, Shimada N, Torihata M, Kuwahara S (2010) Enantioselective total synthesis of idesolide via NaHCO3-promoted dimerization. Tetrahedron 66:4965–4969
Palo RT (1984) Distribution of birch (Betula spp.), willow (Salix spp.), and poplar (Populus spp.) secondary metabolites and their potential role as chemical defense against herbivores. J Chem Ecol 10:499–520
Pauli GF, Poetsch F, Nahrstedt A (1998) Structure assignment of natural quinic acid derivatives using proton nuclear magnetic resonance techniques. Phytochem Anal 9:177–185
Pentzold S, Zagrobelny M, Rook F, Bak S (2014) How insects overcome two-component plant chemical defence: plant β-glucosidases as the main target for herbivore adaptation. Biol Rev Cambridge Philos Soc 89:531–551
Ruuhola T, Tikkanen O-P, Tahvanainen J (2001) Differences in host use efficiency of larvae of a generalist moth, Operophtera brumata on three chemically divergent Salix species. J Chem Ecol 27:1595–1615
Ruuhola T, Julkunen-Tiitto R, Vainiotalo P (2003) In vitro degradation of willow salicylates. J Chem Ecol 29:1083–1097
Shelomi M, Heckel DG, Pauchet Y (2016) Ancestral gene duplication enabled the evolution of multifunctional cellulases in stick insects (Phasmatodea). Insect Biochem Mol Biol 71:1–11
Smith MT (1985) Quinones as mutagens, carcinogens, and anticancer agents: introduction and overview. J Toxicol Environ Health 16:665–672
Stevenson PC, Anderson JC, Blaney WM, Simmonds MS (1993) Developmental inhibition of Spodoptera litura (fab.) larvae by a novel caffeoylquinic acid from the wild groundnut, Arachis paraguariensis (Chod et Hassl.) J Chem Ecol 19:2917–2933
Thieme H (1964) Isolierung eines neuen Phenolglucosids aus Salix purpurea L. Pharmazie 19:725
Wan C, Han J, Chen C, Yao L, Chen J, Yuan T (2016) Monosubstituted benzene derivatives from fruits of Ficus hirta and their antifungal activity against phytopathogen Penicillium italicum. J Agric Food Chem 64:5621–5624
Wang L, Qu L, Zhang L, Hu J, Tang F, Lu M (2016) Metabolic responses of poplar to Apripona germari (hope) as revealed by metabolite profiling. Int J Mol Sci 17:923
Acknowledgements
Open access funding provided by Max Planck Society. The authors thank the workshop and IT-teams of the Max Planck Institute for Chemical Ecology for constructive cooperation, the greenhouse team for rearing the Populus beaupré trees, Regina Seibt for establishing the C. vinula breeding at our institute and Emily Wheeler for polishing the language.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 7545 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Feistel, F., Paetz, C., Menezes, R.C. et al. Acylated Quinic Acids Are the Main Salicortin Metabolites in the Lepidopteran Specialist Herbivore Cerura vinula. J Chem Ecol 44, 497–509 (2018). https://doi.org/10.1007/s10886-018-0945-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10886-018-0945-1