Abstract
Associations of salicylate-like phenolic glycosides (PGs) with biological activity have been reported in Salix and Populus trees, but only for a few compounds, and in relation to a limited number of herbivores. By considering the full diversity of PGs, we may improve our ability to recognize genotypes or chemotype groups and enhance our understanding of their ecological function. Here, we present a fast and efficient general method for salicylate determination in leaves of Eurasian aspen that uses ultra-high performance liquid chromatography-electrospray ionization/time-of-flight mass spectrometry (UHPLC-ESI/TOFMS). The time required for the liquid chromatography separations was 13.5 min per sample, compared to around 60 min per sample for most HPLC protocols. In leaf samples from identical P. tremula genotypes with diverse propagation and treatment histories, we identified nine PGs. We found the compound-specific mass chromatograms to be more informative than the UV-visible chromatograms for compound identification and when quantitating samples with large variability in PG content. Signature compounds previously reported for P. tremoloides (tremulacin, tremuloidin, salicin, and salicortin) always were present, and five PGs (2'-O-cinnamoyl-salicortin, 2'-O-acetyl-salicortin, 2'-O-acetyl-salicin, acetyl-tremulacin, and salicyloyl-salicin) were detected for the first time in P. tremula. By using information about the formic acid adduct that appeared for PGs in the LTQ-Orbitrap MS environment, novel compounds like acetyl-tremulacin could be tentatively identified without the use of standards. The novel PGs were consistently either present in genotypes regardless of propagation and damage treatment or were not detectable. In some genotypes, concentrations of 2'-O-acetyl-salicortin and 2'-O-cinnamoyl-salicortin were similar to levels of biologically active PGs in other Salicaceous trees. Our study suggests that we may expect a wide variation in PG content in aspen populations which is of interest both for studies of interactions with herbivores and for mapping population structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phenolics are bioactive compounds in plants that play diverse roles including structural support of the cell wall, protection from UV irradiation, and defense from herbivory. These compounds derive from distinct branches of the shikimate-phenylpropanoid pathway and include the lignins, flavonoids, anthocyanins, salicylate-like phenolic glycosides (PGs), and tannins. The biosynthesis of those phenolic classes that derive from p-coumaric acid are particularly well described and include monolignols (Sterky et al., 1998; Boerjan et al., 2003), flavonoids (Tsai et al., 2006; Chen et al., 2009), and condensed tannins (Tanner et al., 2003; Dixon et al., 2005). While PGs are suggested to branch from the shikimate-phenylpropanoid pathway “upstream” of p-coumaric acid at either chorismate or cinnamate, their biosynthesis remains poorly understood (Tsai et al., 2006; Morse et al., 2007; Chen et al., 2009; Babst et al., 2010).
Together with condensed tannins, PGs shape resistance against herbivores in the Salicaceous family, including trees of the genera Populus and Salix (Lindroth and Hwang, 1996; Orians et al., 1997; Albrectsen et al., 2007). In general, anti-insect toxicity appears to increase with the complexity of the PGs, in terms of their molecular weight and number of ring structures (Philippe and Bohlman, 2007).
PG profiles vary qualitatively among species but vary quantitatively within species (Palo, 1984; Julkunen-Tiitto, 1986; Greenaway et al., 1991; Orians et al., 1997). Aspen clones can be recognized by their relative PG contents across environments (Lindroth and Hwang, 1996; Mutikainen et al., 2000; Osier and Lindroth, 2006; Tsai et al., 2006; Donaldson and Lindroth, 2007; Chen et al., 2009). When compared to the entire metabolome, which appears to be both dynamic and diverse (Robinson et al., 2005, 2007), PGs are promising compounds for genotype recognition.
Eurasian aspen (P. tremula) is distributed widely across the Northern Hemisphere, and the European part of the population is characterized by a complex genetic structure (De Carvalho et al., 2010). In the Swedish aspen population (SwAsp) of 116 genotypes, this diversity is expressed by phenological variation in bud set and growth differences that mainly follow a latitudinal cline (Luquez et al., 2008). The latitudinal cline also is partly reflected in the distribution of the most abundant and persistent antagonists, rust (Melampsora sp.) and a mining moth (Phyllocnistis labyrinthella) (Albrectsen et al., 2010). However, the interaction between P. tremula and herbivores is poorly understood compared to that of P. tremuloides and its antagonists.
The description of PGs for Eurasian aspen has so far been built on information and methods developed for P. tremuloides, assuming similar PG profiles (Albrectsen et al., 2010). Advanced methods for chemical profiling increase our ability to explore the diversity and dynamics of defense compounds in plants. The purpose of the present study was to examine the PG profiles of P. tremula leaves, using liquid chromatography-mass spectrometry (LC-MS) in order to identify and quantify phenolic glycosides that occur at concentrations indicative of bioactivity. In addition, the study was designed to assess the reliability of this class of phenolic compounds as genotype indicators, and to evaluate their potential as markers to use in future studies of genetic structure and association with resistance traits as assessed through field surveys.
Methods and Materials
Chemicals
Salicortin, salicyloyl-salicin, and tremuloidin standards were gifts from S.D. Mansfield (University of British Columbia, Canada), and 2'-O-cinnamoyl salicortin from C. Orians (Tufts University, MA, USA). Tremulacin and salicin were purchased from Sigma (St. Louis, MO, USA). We follow the nomenclature of Zapesochnaya et al. (2002), in which salicyloyl-salicin corresponds to salicoylsalicin according to Kammerer et al. (2005).
Plant Materials and Treatments
All plants were Populus tremula L. genotypes from the SwAsp Collection (Luquez et al., 2008). The plants had various propagation and treatment histories since one aim of the analyses was to evaluate the robustness of the phenolic glycosides as genotype-specific markers.
For initial characterization and precision analyses, plants representing SwAsp genotypes 7, 13, 47, and 115 were selected. Clones of these genotypes were propagated directly from roots excavated in 2004. The plants were grown in a greenhouse in Umeå, Sweden under ambient conditions since 2005, and cut back so they were about 1 m in height in the fall of 2006. Bud flush occurred in March 2007. In May, when the trees were about 2 m tall, 10 mature leaves were harvested along a 1 m stem section from 2 trees representing each genotype. The harvested leaves were quick-frozen in liquid nitrogen, and stored at −80°C until LC-MS analysis.
Initial Genotypic Profiling
For initial assessment of between-genotype differences in phenolic glycoside profiles, we used leaf material from SwAsp genotypes 5, 23, 36, 47, and 72 propagated as described above. We analyzed two biological replicates of each genotype and two technical replicates per plant.
Reliability of PGs for Genotyping
To assess the reliability of phenolic glycosides as genotypic markers, we compared their profiles in damaged and undamaged plants. The plants used in this experiment included representatives of SwAsp genotypes 5, 23, 36, 47, 51, and 72, which had been propagated in MS tissue culture medium (Murashige and Skoog, 1962) since May 2007. After transfer to the greenhouse, and a month of acclimatization, on November 9th 2007 (at the height of 55.3 ± 10.8 cm (SD), and with 15.8 ± 3.2 leaves per plant), plants were randomly assigned to three treatments: simulated chewing damage; damage by aphids; and controls.
The aphid used in the experiment was Chaitophorus populeti (Panzer). In early September, individuals were collected on Umeå University campus from Populus tremula, where they aggregated with two other Chaitophorus species, probably C. pentandrinus (Ossiannilsson) and C. vitellinae (Schrank). Species were determined by Dr. Richa Raizada. Prior to the experiment, cultures of C. populeti were kept on aspen plants in the greenhouse where they rapidly developed and reproduced.
In Populus, orthostichous leaf positions are 5/10, 6/11 (Jones et al., 1993; Orians and Jones, 2001), i.e., vascular connections are shared between leaf positions 5 and 10, 6 and 11, and so on, with the first fully expanded leaf denoted as ‘leaf 1’. The treatments involved punching multiple minute holes into leaves 11–16 (simulated damage treatment), or placing one reproductive female C. populeti per leaf (position 11–16, aphid treatment). After 96 h, six mature leaves above the experimental leaves (5–10) were harvested as described above. There were 2–3 biological replicates per genotype and treatment, and one technical replicate per plant (48 plants and samples in total).
Extraction
Leaves were ground in a mortar kept frozen using liquid nitrogen, and the frozen powder from each sample (10–12 mg) was extracted by adding it to 1.00 ml of cold chloroform:methanol:H2O (1:3:1) containing 7 ng/μl of [2H6]salicylic acid (Isotec, Miamisburg, OH, USA) as an internal standard. The mixture was agitated with a 3 mm tungsten carbide bead in a MM 301 Vibration Mill (Retsch GmbH and Co. KG, Haan, Germany) at 30 Hz for 3 min (Gullberg et al., 2004). To prevent contamination with tissue debris, extracts were centrifuged at 20,800 × g for 10 min at 4°C, and 200 μl of the supernatant were evaporated to dryness. The residue was dissolved in 10 μl cold methanol and diluted with 90 μl cold water containing 0.1% formic acid, prior to ultra high performance liquid chromatography (UHPLC)-electrospray ionization/time-of-flight mass spectrometry (ESI/TOFMS). A total sample extraction time of less than 2 h at 4°C, from thawing of leaf tissue to dried extract, was used to control decomposition of phenolic glycosides in the samples. Storage of solutions of the standard compounds at 4°C for this time period did not result in any decomposition of PGs.
UHPLC-TOFMS Analysis
The PGs were analyzed by UHPLC-TOFMS using an Acquity™ system, equipped with a column oven (held at 40°C), a 2.1 × 100 mm, 1.7 μm C18 UPLC™ column and an Acquity photodiode array (PDA) detector coupled to an LCT Premier TOFMS (all from Waters, Milford, MA USA). The PGs in a 2 μl injection of each sample were separated with a mobile phase program consisting of 1–20% B over 0–4 min, 20–40% B (4–6 min), 40–95% B (6–9 min), 95% B (9–13.5 min). Solvent B was acetonitrile containing 0.1% formic acid, solvent A was H2O containing 0.1% formic acid, and the flow rate was 500 μl min−1. The eluate was passed through the PDA detector for scanning from 210 to 400 nm, at a sampling rate of 20 points s−1 and resolution of 1.2 nm, and then directly into the ESI source. The source temperature was 120°C, cone gas flow 10 l h−1, desolvation temperature 320°C, nebulization gas flow 600 l h−1, and the capillary and cone voltages were set at 2.5 kV (negative ionization mode) and 35 V, respectively. Data were acquired, in dynamic range enhancement (DRE) mode, every 0.1 s, with a 0.01 s interscan delay. Leucine enkephalin was the lock mass compound for accurate mass measurements, infused directly at 400 pg μl−1 in 50:50 acetonitrile:H2O at 20 μl min−1. The normal lock mass in DRE mode was the negative 13 C ion of leucine enkephalin (m/z 555.2654), and the extended lock mass was the normal negative ion at m/z 554.2620. Mass spectra were acquired in centroid mode, m/z 100–1000, with a data threshold value set to 3. In a complementary experiment, in-source fragmentation was increased by increasing the aperture 1 voltage from 5 to 45 V to exclude adduct ion formation.
Profiling–Data Processing
The UHPLC-ESI/TOFMS instruments were operated with MassLynx™ v. 4.1 software (Waters, Milford, MA, USA), and the QuanLynx module of this package was used to assess relative amounts of phenolic glycosides by calculating peak areas from both mass chromatograms (see Table 1, ESI-MS m/z with 0.10 Da mass windows), and specific wavelength chromatograms using the compound-specific UV λmax values (Table 1). The peak areas were normalized against that of the internal standard salicylic acid (M = [C7H 22 H4O3], RT = 5.01 min, m/z 141.0495, 187.0550, and 305.0883 for the [M-H]−, [M-H + HCOOH]− and [M2−2H + Na]− ions, respectively; λmax 231 and 305 nm), as well as the mass of tissue used for extraction.
Phenolic Glycoside Structural Identification (UHPLC-LTQ/Orbitrap)
To elucidate the structures of the compounds, they were separated by a Thermo Accela LC system, equipped with a column oven (held at 40°C) and a Hypersil C18 GOLD™ column (2.1 × 50 mm, 1.9 μm; mobile phase as for the UHPLC-ESI/TOFMS) and analyzed by tandem mass spectrometry using a LTQ/Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Centroid mass spectra of negative ions were collected in the Orbitrap mass analyzer, with a target mass resolution of 30,000 (full width at half maximum peak height, defined at m/z 400), after collision-induced dissociation (CID) in the LTQ cell, using a normalized collision energy of 20%. External mass calibration was performed according to the manufacturer’s guidelines.
Analytical Precision
To assess the precision of the analyses, 5 samples of leaves of each of 2 genotypes (7 and 115) were extracted, and part of each extract was analyzed in parallel, while the other parts were pooled by genotype. To test for methodical errors, the pooled extracts were each divided into 10 microvials, and analyzed by UHPLC-PDA-ESI/TOFMS. In addition, 5 repeat injections from 1 individual microvial were performed to determine injection and instrumentation errors. To check for retention time and mass accuracy throughout the experiment, a standard mix consisting of 5 standards (salicin, chlorogenic acid, rutin, nalidixic acid, and phloretin), each at 50 μM in 50% acetonitrile, was injected after every ten injections.
Concentration Determination
Standards of various concentrations (0.5 ng–10 μg injected) were analyzed by UHPLC-PDA-ESI/TOFMS as described above, and the peak areas from the resulting mass chromatograms were used to construct calibration curves.
Statistical Analysis
Multivariate analysis of PG profiles was performed using principal component analysis (PCA), with unit variance scaling, in SIMCA-P+ v. 12.0 (Umetrics AB, Umeå, Sweden). Relative standard deviations (RSD) were measured for reproducibility and RSD in the Umetrics software equals the coefficient of variation (CV).
Results
Identification
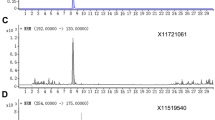
With LC-ESI/TOFMS operating in negative ionization mode, we identified nine salicylate-like phenolic glycosides in P. tremula leaves: salicin, salicortin, tremuloidin, tremulacin, 2'-O-acetyl-salicin, 2'-O-acetyl-salicortin, salicyloyl-salicin, 2'-O-cinnamoyl-salicortin, and acetyl-tremulacin (Fig. 1, Table 1). The identifications were based on comparisons of the LC-retention times, UV spectra, and accurate masses of the compounds detected in the extracts with those of standards (Table 1). These nine phenolic glycosides were baseline-separated by the LC (Fig. 2a), and they constituted some of the most intense peaks in the total ion chromatograms, suggesting that they occurred in high concentrations (Fig. 2b).
UHPLC-PDA-ESI/TOFMS analysis of a plant extract (genotype 47). a Single mass chromatograms (m/z window 0.04) for the nine phenolic glycosides. b Total ion chromatogram showing relative intensities of peaks from TOFMS, with the mass spectrum for tremulacin inserted. c Full wavelength chromatogram from the PDA detector (UV, 210–400 nm) with the absorbance spectrum for tremulacin (peak at 6.45 min) inserted. Since the PDA detector was connected between the LC-column and the TOFMS, the retention time was ca. 0.3 min shorter than in MS detection. Asterisks indicate peaks corresponding to phenolic glycosides. The LC-retention times are indicated
Phenolic glycosides are aromatic, and thus can be detected by monitoring UV absorbance. The summed PDA absorbance (UV) chromatograms (Fig. 2c) had fewer peaks than the total ion chromatograms acquired by TOFMS (Fig. 2b). However, the peaks that were detected displayed characteristic absorbance spectra of phenolic glycosides, with two UV peaks (Fig. 2c insert and Table 1), one at about 215–220 nm (but 240 nm for salicyloyl-salicin), and a weaker one at 270 nm. Either wavelength could be used for quantification (Table 2).
A drawback of using ESI-MS is the formation of adducts. With the ESI/TOFMS instrument used in these analysis, adducts with acetonitrile, formic acid and/or sodium ion have been detected in the negative ionization mode, and ammonium adducts found in the positive mode. For all of the phenolic glycosides, the mass spectra showed a base peak of the deprotonated molecular ion as a formic acid adduct [M-H + HCOOH]−, presumably arising from combination with the formic acid present in the LC mobile phase. The intensity of the unmodified molecular ion was very low (Fig. 2b insert and Table 1) for the PGs, except for salicyloyl-salicin in the samples as well as in pure standards. In the complementary analysis with increased in-source dissociation in the ESI/TOFMS, the deprotonated molecular ion and the formic acid adduct were both present, confirming the phenolic glycoside masses.
The initial identifications of the three novel acetylated phenolic glycosides were based on their masses and absorbance spectra. To verify their identities, the extracts also were analyzed by LC-MS/MS using an LTQ/Orbitrap system (Hu et al., 2005; Makarov et al., 2006) (Fig. 3, Suppl. Fig. S1) and structural identification of the novel phenolic glycosides was obtained from the fragmentation patterns. The ESI system of this instrument also generated formic acid adducts, but it yielded deprotonated, unmodified ions at higher intensities, and the latter were preferentially selected by mass for the MS/MS analysis. Utilizing the high mass accuracy of the Orbitrap MS, the elemental compositions of these phenolic glycosides confirmed that they consisted of acetylated versions of salicin, salicortin, and tremulacin. The acetyl-salicortin (Fig. 3b) fragments were similar to those from salicortin (Suppl. Fig. S1d; Kammerer et al., 2005), with several fragments that increased by 42 Da in the acetyl-salicortin spectrum. Therefore, we concluded that the acetyl group was attached to the glucose moiety of the salicortin. Similar conclusions were drawn for acetyl-salicin (Fig. 3a) and acetyl-tremulacin (Fig. 3e). The characteristics of the UV spectra, as well as the slightly longer retention times compared to those of the non-acetylated phenolic glycosides (Table 1), corroborated our structural assignment. We suggest that the acetyl group of acetyl-tremulacin is bonded to the 6'-O of the glucose ring, since this seems to be the second most reactive position, as in diacetyl-salicortin (Ruuhola et al., 2003; Ruuhola and Julkunen-Tiitto, 2003). We propose that the 2'-O position is occupied by the benzoyl moiety in tremulacin, and that the acetyl groups of acetyl-salicin and acetyl-salicortin occupy the 2’-O-positions of salicin and salicortin, as found in willows (Ruuhola and Julkunen-Tiitto, 2000).
CID MS/MS spectra of the newly identified phenolic glycosides: a 2'-O-acetyl-salicin, b 2'-O-acetyl-salicortin, c salicyloyl-salicin, d 2'-cinnamoyl-salicortin, and e acetyl-tremulacin. Respective m/z selected for fragmentation is indicated. Compound structures are shown with proposed bond breakages during fragmentations
Method Reproducibility
To validate the UHPLC-PDA-ESI/TOFMS method for analyzing phenolic glycosides we used it to analyze multiple sub-samples from the same (pooled) extract. The reproducibility was within acceptable ranges for detectable peaks with RSDs, of 4–7% for UV detection, and 6–12% for MS detection (Table 2). Sequential injections from the same microvial showed little variation, with RSDs as low as <2% for UV, and 1–5% for MS quantification (Table 2), indicating little instrumental variation. Parallel extractions from subsamples of the same plant tissue were performed to explore the reproducibility of all steps in the analysis, and resulted in somewhat higher variations (Table 2).
Phenolic Glycoside Concentrations
Six standards (salicin, salicortin, tremuloidin, salicyloyl-salicin, tremulacin, and 2'-O-cinnamoyl-salicortin) at various concentrations were analyzed by UHPLC-PDA-ESI/TOFMS, and the calibration curves obtained from the resulting mass chromatograms were used for quantification. The UV and MS detection limits for individual phenolic glycosides varied (Table 3), but mass spectrometry could generally detect lower concentrations. Therefore, UV detection may not be optimal when screening several genotypes with highly variable phenolic glycoside concentrations. In such cases, MS detection may be more suitable for quantification, at the expense of somewhat lower reproducibility (Table 2).
Phenolic glycosides concentrations were calculated per unit mass of leaf powder (Table 3). Salicin was found at 0.5–2.4 μg mg−1 and tremulacin at 0.1–4.0 μg mg−1, depending on the genotype. These levels are comparable to concentrations found in P. tremuloides (Lindroth and Hwang, 1996; Robinson et al., 2005; Harding et al., 2009). In some genotypes, 2'-O-cinnamoyl-salicortin was detected in concentrations up to 2.1 μg mg−1, which is within the range of 0.75–5.6 μg mg−1 found in Salix sericea (Orians and Fritz, 1995). In other genotypes, this compound was below the detection limit of about 0.25 μg/mg tissue. Salicortin, tremuloidin, and salicyloyl-salicin were found within ranges of 1.8–6.9, 0.1–1.1, and 0–0.12 μg mg−1, respectively. Preliminary calculations based on the assumption that concentration/signal intensity ratios were similar to the corresponding unacetylated compounds suggest that acetyl-salicin, acetyl-salicortin, and acetyl-tremulacin were present at concentrations ranging from 0 to 1.9, 0 to 2.9, and 0 to 0.2 μg mg−1, respectively.
Phenolic Glycosides as Genotype-Specific Markers
PCA of the data acquired in the initial assessment of between-genotype differences in phenolic glycoside profiles suggests that the five tested genotypes clustered into three distinct chemotype-groups (score plot, Fig. 4a and Fig. S3) and that the nine phenolic glycosides all contributed to this pattern (loadings, Fig. 4a). The PCA model showed two significant components, explaining 81% of the variation in the data [R 2 X(cum) = 0.812; Q 2(cum) = 0.565]. Variations in contents of 2'-O-cinnamoyl-salicortin, the acetylated phenolic glycosides, and to some extent tremulacin and tremuloidin were largely responsible for separating the genotypes along the first principal component axis (PC1).
Multivariate analysis of phenolic glycoside profiles of Populus tremula genotypes from the SwAsp collection. a PCA score plot (PC1 versus PC2) of data acquired from analyses of samples of greenhouse-grown plants representing five genotypes (5, 23, 36, 47, and 72), with data marked by genotype, and the corresponding loading plot. b Corresponding plots of data acquired from analyses of plants representing six genotypes (5, 23, 36, 47, 51, and 72) exposed to three treatments: simulated damage (= wounded), aphid infestation and controls, with data coded by (I) genotype, and (II) treatment. Solid circles show 95% Hotelling T 2 confidence regions, and dashed circles indicate chemotype-groups (in a, b-I), and treatment groups (in b-II)
Analysis of the phenolic glycoside profiles of representatives of an overlapping set of genotypes, following varying propagation and stress treatments, suggested a similar pattern; six genotypes formed two distinct clusters or chemotype-groups (Fig. 4b-I). One chemotype-group included three separate genotypes (47, 51, and 72) that contained acetylated phenolic glycosides, while the other included three genotypes (5, 23, 36) characterized by the presence of 2'-O-cinnamoyl-salicortin (Fig. 4b-I). Again, all phenolic glycosides contributed to the clustering of the genotypes (loadings, Fig. 4b), although 2'-O-cinnamoyl-salicortin had the heaviest loading on the first axis (PC1). Since PCA did not detect any treatment effects (Fig. 4b-II), the phenolic glycosides appear to be potentially robust chemotype group markers and, to some extent genotype markers in P. tremula. The PCA model constructed from the data acquired from the analyses of samples of plants subjected to various treatments had four significant principal components, which together explained 96% of the variation in the data [R 2 X(cum) = 0.955; Q 2(cum) = 0.831].
The consistent separation into chemotype-groups in the two data sets (Fig. 4a and b) was confirmed by studies of separate profiles for each plant and phenolic glycoside compound (Suppl. Fig. S2, showing normalized data obtained in the PG consistency analysis). Depending on the genotype, the phenolic glycosides 2'-O-acetyl-salicin, 2'-O-acetyl-salicortin, 2'-O-cinnamoyl-salicortin, and acetyl-tremulacin, were either consistently present in substantial amounts, or consistently below the detection limit. The structural similarities of the compounds that contributed significantly to the patterns detected in the multivariate analyses are indicated in the loading plot (Suppl. Fig. S3).
Discussion
Nine phenolic glycosides were found by UHPLC-ESI/TOFMS in extracts of European aspen (P. tremula) leaves. Salicin, salicortin, tremuloidin, and tremulacin were present in all tested genotypes, in agreement with the “signature” phenolic glycosides reported for P. tremuloides (Lindroth and Hwang, 1996; Harding et al., 2009). Salicyloyl-salicin also was found in most extracts. In addition, some genotypes contained considerable amounts of 2'-O-acetyl-salicin, 2'-O-acetyl-salicortin, 2'-O-cinnamoyl-salicortin, and/or acetyl-tremulacin, which have not been described previously in either P. tremula or P. tremuloides.
Strengths of UHPLC-MS
A TOFMS instrument is suitable for identifying phenolics, when standard compounds are available, but it has limited utility for identifying novel compounds, since structural elucidations require MS/MS analysis for identification and confirmation. For this purpose, the high mass-accuracy LTQ/Orbitrap instrument was used here, although other MS/MS compatible instruments could have been used with equally good results. Selected reaction monitoring (SRM) methods using an instrument such as triple quadrupole (QQQ) might also have been used for quantification purposes, but would require prior knowledge about compound masses. Well established HPLC protocols take 40 to 60 min per sample to analyze phenolic glycosides (Ruuhola et al., 2003; Ruuhola and Julkunen-Tiitto, 2003; Warren et al., 2003). However, by using ultra high performance liquid chromatography (UHPLC), the analyses are faster and the resolution is better, resulting in higher precision. The methodology for phenolic glycosides presented here reduced the time required for the liquid chromatography separations to just 13.5 min.
Our results also showed the limitation of detection by UV, in comparison to MS. Although UV detection generally provided the best reproducibility for individual phenolic glycosides, it is less sensitive than MS for phenolic glycosides. When large numbers of samples with variable concentrations are to be analyzed, MS measurements are more suitable, since they can cope with a wider range of concentrations. In addition, an extracted mass chromatogram is more compound specific than an extracted UV-visible chromatogram; in the latter, contributions from coeluting compounds are harder to distinguish and might affect the quantification. Hence, the TOF and Orbitrap MS analysis was essential for characterizing the phenolic glycosides in the aspen leaf extracts. Depending on the objective, less powerful MS systems might be sufficient, although a rapidly scanning instrument is highly advantageous for collecting many data points over each chromatographic elution peak. While in recent years hundreds of UHPLC-base methods have been published for analyzing various classes of metabolites, this is the first method for phenolic glycoside analysis to date.
Can Phenolic Glycoside Profiles Be Used to Characterise Populus-Genotypes?
Multivariate (PCA) analysis of the PG profiles clustered the genotypes into similar groups, regardless of the treatments applied (Fig. 4). Hence, the phenolic glycosides provided a means for recognizing chemotype groups of P. tremula across treatments. Salicin, salicortin, tremuloidin, and tremulacin were present in all samples and varied with genotype. These four compounds are well-known to vary quantitatively with P. tremuloides genotype (Lindroth and Hwang, 1996; Harding et al., 2009). In contrast, the five novel phenolic glycosides in P. tremula followed a discrete pattern of presence or absence in a given genotype. Moreover, when present, the novel compounds also varied in quantity in a genotypic specific pattern, and contents of 2'-O-cinnamoyl-salicortin, and the acetylated phenolic glycosides were largely responsible for separating the genotypes along the first principal component axis. (for profiles of particular phenolic glycosides see progeny data provided in Suppl. Fig. S2). The pattern of appearance of these novel phenolic glycosides suggests that they are potential Mendelian traits i.e., traits controlled by a single loci. Such traits are promising for studies of both herbivore resistance and genetic structure. Populations of P. tremula are polymorphic, and thus genetically highly variable for several growth traits (Ingvarsson, 2005; De Carvalho et al., 2010), many of which do not follow simple Mendelian inheritance (Luquez et al., 2008). The novel phenolic glycosides not only confirm that aspens in Sweden may constitute a complex genetic composition, but also offer new traits that may be helpful in understanding the population structure of Eurasian aspen.
Can We Expect to Find Other PGs in P. tremula?
Novel phenolic glycosides were detected by identifying compounds with salicylate-like UV-spectra combined with LC-ESI/TOFMS-analysis. Peaks that had similarities to known phenolic glycosides were analyzed further by LTQ-Orbitrap MS, resulting in accurate mass determination, and tandem mass spectra for structural interpretation. At the population level, there may be more salicylate-like phenolic compounds to discover in P. tremula, and only careful survey of a larger number of genotypes will provide a comprehensive list of compounds. Many salicylate-like phenolic glycosides have been described in Salicaceous species (Ruuhola and Julkunen-Tiitto, 2000, 2003; Kammerer et al., 2005; Tsai et al., 2006), and although we can exclude the possibility that additional PGs were present in the major peaks in the samples that we screened, more genotypes should be screened to obtain information about their diversity at stand and population levels. In the samples examined in this study, the phenolic glycosides always were associated with formic acid under our LC-MS conditions, which led to the detection of acetyl-tremulacin. Thus, assuming that an acetylated tremuloidin compound would also associate with formic acid, we specifically searched for such a compound (using m/z 431.1348 and 477.1402 for the compound with and without a formic acid adduct) and concluded it was not present in our samples in detectable amounts.
Implications of PG Diversity
Biosynthetic routes of the salicylate-like phenolic glycosides have proven less easy to elucidate compared to those for flavonoids (Tsai et al., 2006; Chen et al., 2009; Babst et al., 2010). On the basis of correlated patterns in the loading plots (Fig. 4), the biosynthesis of acetylated phenolic glycosides appear related to that of non-acetylated molecules with similar structures (Fig. 1), thus supporting conclusions drawn by Ruuhola and Julkunen-Tiitto (2003). Moreover, some phenolic glycosides might be metabolically interconverted (Ruuhola and Julkunen-Tiitto, 2003; Babst et al., 2010), a possibility which would agree with the close correlation between salicin and salicortin in our loading plots. Salicin and salicortin appear on the second principal component axis in the PCA plots, rather than the first component, which is responsible for the main separation among genotypes, providing further support for a dynamic interrelationship between salicin and salicortin.
Phenolic glycosides have well-established biological activities and utility for distinguishing among genotypes within and among species. Salicortin and tremulacin, for example, are associated with resistance to defoliating insect herbivores in P. tremuloides (Hemming and Lindroth, 1995; Osier and Lindroth, 2006; Donaldson and Lindroth, 2007; Wimp et al., 2007), while salicortin and 2'-O-cinnamoyl-salicortin are associated with resistance to a range of herbivores within the North American Salix sericea × eriocephala hybrid complex (Orians et al., 1997). Diverse phenolic glycoside composition also has been identified in Finnish and Swedish Salix spp., including 2'-O-acetyl-salicin and 2'-O-acetyl-salicortin in S. pentandra (Ruuhola and Julkunen-Tiitto, 2003); although in these species, the relationships between specific compounds and biological activities appear to be complex or non-existent, thus highlighting the ambiguous ecological functions of phenolic glycosides (Hjältén et al., 2007; Witzell and Martin, 2008; Albrectsen et al., 2010).
Compared to other Salicaceous study systems, we have little information regarding biological activity of phenolic glycosides in P. tremula. Generally, the intra-specific diversity expressed in the modest number of genotypes in the present study suggests that the phenolic glycoside composition of P. tremula is more complex than that of P. tremuloides. Given the great differences in damage regimes in the two aspen species and the defensive role that phenolic glycosides often play against various pests, the differences in distribution of defense compounds, warrant further attention. Hence, a detailed comparative survey of phenolic glycoside diversity at population level in North American and Eurasian aspen may help to elucidate their ecological function.
References
Albrectsen, B. R., Gutierrez, L., Fritz, R. S., Fritz, R. D., and Orians, C. M. 2007. Does the differential seedling mortality caused by slugs alter the foliar traits and subsequent susceptibility of hybrid willows to a generalist herbivore? Ecol. Entomol. 32:211–220.
Albrectsen, B. R., Witzell, J., Robinson, K. M., Wulff, S., Luquez, V. M. C., Agren, R., and Jansson, S. 2010. Large scale geographic clines of parasite damage to Populus tremula L. Ecography. 33:483–493.
Babst, B. A., Harding, S. A., and Tsai, C. J. 2010. Biosynthesis of phenolic glycosides from phenylpropanoid and benzenoid precursors in Populus. J. Chem. Ecol. 36:286–297.
Boerjan, W., Ralph, J., and Baucher, M. 2003. Lignin biosynthesis. Annu. Rev. Plant. Biol. 54:519–546.
Chen, F., Liu, C. J., Tschaplinski, T. J., and Zhao, N. 2009. Genomics of secondary metabolism in Populus: interactions with biotic and abiotic environments. Crit. Rev. Plant. Sci. 28:375–392.
De Carvalho, D., Ingvarsson, P. K., Joseph, J., Suter, L., Sedivy, C., Macaya-Sanz, D., Cottrell, J., Heinze, B., Schanzer, I., and Lexer, C. 2010. Admixture facilitates adaptation from standing variation in the European aspen (Populus tremula L.), a widespread forest tree. Mol. Ecol. 19:1638–1650.
Dixon, R. A., Xie, D. Y., and Sharma, S. B. 2005. Proanthocyanidins—a final frontier in flavonoid research? New. Phytol. 165:9–28.
Donaldson, J. R., and Lindroth, R. L. 2007. Genetics, environment, and their interaction determine efficacy of chemical defense in trembling aspen. Ecology. 88:729–739.
Greenaway, W., English, S., May, J., and Whatley, F. R. 1991. Chemotaxonomy of section leuce poplars by GC-MS of bud exudate. Biochem. Syst. Ecol. 19:507–518.
Gullberg, J., Jonsson, P., Nordstrom, A., Sjostrom, M., and Moritz, T. 2004. Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsis thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal. Biochem. 331:283–295.
Harding, S. A., Jarvie, M. M., Lindroth, R. L., and Tsai, C. J. 2009. A comparative analysis of phenylpropanoid metabolism, N utilization, and carbon partitioning in fast- and slow-growing Populus hybrid clones. J. Exp. Bot. 60:3443–3452.
Hemming, J. D. C., and Lindroth, R. L. 1995. Intraspecific variation in aspen phytochemistry—effects on performance of gypsy moths and forest tent caterpillars. Oecologia. 103:79–88.
Hjältén, J., Niemi, L., Wennstrom, A., Ericson, L., Roininen, H., and Julkunen-Tiitto, R. 2007. Variable responses of natural enemies to Salix triandra phenotypes with different secondary chemistry. Oikos. 116:751–758.
Hu, Q. Z., Noll, R. J., Li, H. Y., Makarov, A., Hardman, M., and Cooks, R. G. 2005. The Orbitrap: a new mass spectrometer. J. Mass Spectrom. 40:430–443.
Ingvarsson, P. K. 2005. Nucleotide polymorphism and linkage disequilbrium within and among natural populations of European Aspen (Populus tremula L., Salicaceae). Genetics. 169:945–953.
Jones, C. G., Hopper, R. F., Coleman, J. S., and Krischik, V. A. 1993. Control of systemically induced herbivore resistance by plant vascular architecture. Oecologia. 93:452–456.
Julkunen-Tiitto, R. 1986. A chemotaxonomic survey of phenolics in leaves of northern Salicaceae species. Phytochemistry. 25:663–667.
Kammerer, B., Kahlich, R., Biegert, C., Gleiter, C. H., and Heide, L. 2005. HPLC-MS/MS analysis of willow bark extracts contained in pharmaceutical preparations. Phytochem. Analysis. 16:470–478.
Lindroth, R. L., and Hwang, S.-Y. 1996. Clonal variation in foliar chemistry of quaking aspen (Populus tremuloides Michx.). Biochem. Syst. Ecol. 24.
Luquez, V., Hall, D., Albrectsen, B. R., Karlsson, J., Ingvarsson, P., and Jansson, S. 2008. Natural phenological variation in aspen (Populus tremula): the SwAsp collection. Tree Genet. Genomes. 4:279–292.
Makarov, A., Denisov, E., Kholomeev, A., Baischun, W., Lange, O., Strupat, K., and Horning, S. 2006. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78:2113–2120.
Morse, A. M., Tschaplinski, T. J., Dervinis, C., Pijut, P. M., Schmelz, E. A., Day, W., and Davis, J. M. 2007. Salicylate and catechol levels are maintained in nahG transgenic poplar. Phytochemistry. 68:2043–2052.
Murashige, T., and Skoog, F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plantarum. 15:473–&.
Mutikainen, P., Walls, M., Ovaska, J., Keinanen, M., Julkunen-Tiitto, R., and Vapaavuori, E. 2000. Herbivore resistance in Betula pendula: Effect of fertilization, defoliation, and plant genotype. Ecology. 81:49–65.
Orians, C. M., and Fritz, R. S. 1995. Secondary chemistry of hybrid and parental willows—phenolic glycosides and condensed tannins in Salix sericea, S. eriocephala, and their hybrids. J. Chem. Ecol. 21:1245–1253.
Orians, C. M., and Jones, C. G. 2001. Plants as resource mosaics: a functional model for predicting patterns of within-plant resource heterogeneity to consumers based on vascular architecture and local environmental stability. Oikos 94:493–504.
Orians, C. M., Huang, C. H., Wild, A., Dorfman, K. A., Zee, P., Dao, M. T. T., and Fritz, R. S. 1997. Willow hybridization differentially affects preference and performance of herbivorous beetles. Entomol. Exp. Appl. 83:285–294.
Osier, T. L., and Lindroth, R. L. 2006. Genotype and environment determine allocation to and costs of resistance in quaking aspen. Oecologia. 148:293–303.
Palo, R. T. 1984. Distribution of birch (Betula spp), willow (Salix spp), and poplar (Populus spp) secondary metabolites and their potential role as chemical defense against herbivores. J Chem. Ecol. 10:499–520.
Philippe, R. N., and Bohlman, J. 2007. Poplar defense against insect herbivores. Can. J. Forest Res. 85:1111–1126.
Robinson, A. R., Gheneim, R., Kozak, R. A., Ellis, D. D., and Mansfield, S. D. 2005. The potential of metabolite profiling as a selection tool for genotype discrimination in Populus. J. Exp. Bot. 56:2807–2819.
Robinson, A. R., Ukrainetz, N. K., Kang, K. Y., and Mansfield, S. D. 2007. Metabolite profiling of Douglas fir (Pseudotsuga menziesii) field trials reveals strong environmental and weak genetic variation. New. Phytol. 174:762–773.
Ruuhola, T., and Julkunen-Tiitto, R. 2000. Salicylates of intact Salix myrsinifolia plantlets do not undergo rapid metabolic turnover. Plant Physiology. 122:895–905.
Ruuhola, T., and Julkunen-Tiitto, R. 2003. Trade-off between synthesis of salicylates and growth of micropropagated Salix pentandra. J. Chem. Ecol. 29:1565–1588.
Ruuhola, T., Julkunen-Tiitto, R., and Vainiotalo, P. 2003. In vitro degradation of willow salicylates. J. Chem. Ecol. 29:1083–1097.
Sterky, F., Regan, S., Karlsson, J., Hertzberg, M., Rohde, A., Holmberg, A., Amini, B., Bhalerao, R., Larsson, M., Villarroel, R., et al. 1998. Gene discovery in the wood-forming tissues of poplar: Analysis of 5,692 expressed sequence tags. Proc. Natl. Acad. Sci. USA. 95:13330–13335.
Tanner, G. J., Francki, K. T., Abrahams, S., Watson, J. M., Larkin, P. J., and Ashton, A. R. 2003. Proanthocyanidin biosynthesis in plants—Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol. Chem. 278:31647–31656.
Tsai, C. J., Harding, S. A., Tschaplinski, T. J., Lindroth, R. L., and Yuan, Y. N. 2006. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New. Phytol. 172:47–62.
Warren, J. M., Bassman, J. H., Fellman, J. K., Mattinson, D. S., and Eigenbrode, S. 2003. Ultraviolet-B radiation alters phenolic salicylate and flavonoid composition of Populus trichocarpa leaves. Tree Physiol. 23:527–535.
Wimp, G. M., Wooley, S., Bangert, R. K., Young, W. P., Martinsen, G. D., Keim, P., Rehill, B., Lindroth, R. L., and Whitham, T. G. 2007. Plant genetics predicts intra-annual variation in phytochemistry and arthropod community structure. Mol. Ecol. 16:5057–5069.
Witzell, J., and Martin, J. A. 2008. Phenolic metabolites in the resistance of northern forest trees to pathogens—past experiences and future prospects. Can. J. Forest. Res. 38:2711–2727.
Zapesochnaya, G. G., Kurkin, V. A., Braslavskii, V. B., and Filatova, N. V. 2002. Phenolic compounds of Salix acutifolia bark. Chem. Nat. Compd. 38:314–318.
Acknowledgements
This study was supported by the Carl Tryggers Foundations, VR/Vinnova, and SSF (BA), and KAW, FORMAS, the Kempe Foundation and VR/Vinnova (TM). We thank Drs Richard Lindroth, Colin Orians, Ken Keefover-Ring, and anonymous reviewers for their valuable comments, which greatly improved the manuscript. We also thank Dr. Rajarshi Kumar Gaur for his help with the aphid experiment.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ilka Nacif Abreu and Maria Ahnlund contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
CID MS/MS spectra of phenolic glycosides: a salicin, b salicortin, c tremuloidin, and d tremulacin. Respective m/z selected for fragmentation is indicated. Compound structures are shown with proposed bond breakages during fragmentations. (PDF 90.6 kb)
Supplementary Figure S2
Individual phenolic glycoside profiles for the 48 plants used in the damage experiment, grouped by genotype (5, 23, 36, 47, 51, and 72) and damage treatment. Relative concentrations (%) of the phenolic glycosides measured after three treatments—aphid infestation (A), controls (C), and simulated damage (S)—are shown. (PDF 612 kb)
Supplementary Figure S3
Loading plot from PCA analysis of the data acquired in the initial genotypic profiling with compound structures (as shown in Fig. 4a-loading). (PDF 283 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Abreu, I.N., Ahnlund, M., Moritz, T. et al. UHPLC-ESI/TOFMS Determination of Salicylate-like Phenolic Gycosides in Populus tremula Leaves. J Chem Ecol 37, 857–870 (2011). https://doi.org/10.1007/s10886-011-9991-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10886-011-9991-7