Abstract
The multi-subunit vacuolar-type H+-ATPase consists of a V1 domain (A–H subunits) catalyzing ATP hydrolysis and a V0 domain (a, c, c′, c″, d, e) responsible for H+ translocation. The mammalian V0 d subunit is one of the least-well characterized, and its function and position within the pump are still unclear. It has two different forms encoded by separate genes, d1 being ubiquitous while d2 is predominantly expressed at the cell surface in kidney and osteoclast. To determine whether it forms part of the pump’s central stalk as suggested by bacterial A-ATPase studies, or is peripheral as hypothesized from a yeast model, we investigated both human d subunit isoforms. In silico structural modelling demonstrated that human d1 and d2 are structural orthologues of bacterial subunit C, despite poor sequence identity. Expression studies of d1 and d2 showed that each can pull down the central stalk’s D and F subunits from human kidney membrane, and in vitro studies using D and F further showed that the interactions between these proteins and the d subunit is direct. These data indicate that the d subunit in man is centrally located within the pump and is thus important in its rotary mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The multi-subunit vacuolar-type proton pump (H+- or V-ATPase) is essential for acidification of diverse intracellular compartments in eukaryotic cells, including endosomes, lysosomes, clathrin-coated and synaptic vesicles, chromaffin granules and the central vacuoles of plants and fungi (Forgac 2007). In higher organisms H+-ATPases also function to pump protons across the plasma membrane of certain specialized cell types. These plasma membrane H+-ATPases are involved in processes such as urinary acidification and the maintenance of acid-base homeostasis (Wagner et al. 2004), bone resorption by osteoclasts (Blair et al. 1989), sperm maturation and viability (Pastor-Soler et al. 2005), and the invasiveness of some tumour cells and vascular endothelial cells (Sennoune and Martinez-Zaguilan 2007). H+-ATPases thus represent potential drug targets in the treatment of diseases such as osteoporosis and cancer.
H+-ATPases consist of two functional sectors, V1 and V0. The peripheral V1 domain (subunits A-H) binds and hydrolyzes ATP, providing the energy for proton translocation across the integral membrane V0 domain (subunits a, c, c′ c″, d and e). Whether the c′ subunit is found in species other than yeast is currently unclear. H+-ATPases have been shown to function via a rotary mechanism (Hirata et al. 2003; Imamura et al. 2003). ATP hydrolysis is carried out by a hexamer of V1 subunits A and B. Energy from this reaction drives the rotation of a central stalk consisting of V1 subunits D and F and this is coupled to rotation of the V0 proteolipid ring made up of c, c′ and c″. Proton translocation across the membrane takes place at the interface between the rotating proteolipid ring and the stationary V0 subunit a, which is held fixed in relation to the catalytic head by peripheral stalks which consist of subunits C, E, G and H.
In higher eukaryotes, the B, C, E, G, a, d and e subunits have recently been shown to have multiple forms encoded by different genes and with differing tissue expression patterns (reviewed in Forgac 2007). Importantly, H+-ATPases that function in different physiological processes at different locations appear to contain unique combinations of these subunit isoforms, and thus it may be possible to design isoform-specific inhibitors that could serve as potential therapeutic agents in treating H+-ATPase-related diseases. However for this to be feasible, it is important to understand better the overall structure, subunit composition and function of all the components of the proton pump complex.
The V0 d subunit is one of the least characterized of all the H+-ATPase components, and we have previously shown that in man it exists in two different forms, d1 and d2, encoded by separate genes. These are located on chromosomes 16q22 and 8q21 respectively. The d1 gene is ubiquitously expressed and d2 is predominantly expressed in kidney and osteoclast (Smith et al. 2002). The function of the d subunit and its position within the pump complex are still under debate. It associates with the membrane bound V0 domain, probably by protein-protein interactions rather than being directly anchored in the membrane (Bauerle et al. 1993).
Currently there are two hypotheses regarding its localization. The first is based on studies of subunit C in the closely related bacterial A-ATPase (Fig. 1a). The crystal structure of subunit C in Thermus thermophilus has been solved using two crystal forms, hexagonal (Iwata et al. 2004) and monoclinic (Numoto et al. 2004), which reveal that the protein has an alpha-helical three-domain structure. As a result of this and cross-linking experiments, subunit C is thought to have a socket-like function attaching the central stalk subunits D and F to the proteolipid ring, and although the eukaryotic d subunit has only a low level of sequence identity with bacterial subunit C, it has been proposed to be similar in structure and function (Iwata et al. 2004).
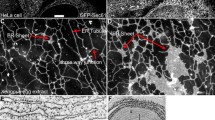
a Schematic diagram of the A-ATPase from T. thermophilus showing A1 and A0 domain subunits (open and hatched symbols respectively) with subunit C indicated in red (adapted from Iwata et al. 2004). b 3D structural model of the H+-ATPase V0 domain from bovine brain clathrin coated vesicles, derived from single particle image analysis of electron micrographs (Wilkens and Forgac 2001), seen from the top (1) and the side (2). The arrows indicate the proposed peripheral position of the boxing glove shaped yeast d subunit (green; Thaker et al. 2007). c Schematic representation of the eukaryotic vacuolar H+-ATPase, showing V1 and V0 domain subunits (open and hatched symbols respectively). In this model, the d subunit (in red) forms part of a peripheral stalk (adapted from Thaker et al. 2007)
The second hypothesis is based on yeast studies and has resulted from the recent determination of the low-resolution structure of the yeast d subunit in solution using small-angle X-ray scattering data (Thaker et al. 2007), which showed the protein to be a boxing glove shaped molecule. The authors of this study have suggested that their structure corresponds to the peripheral density seen partly covering the cytoplasmic opening of the bovine H+-ATPase proton channel in the 3D structural model derived from single particle image analysis of electron micrographs by Wilkens and Forgac (2001; Fig. 1b). Thus the d subunit would be located above and slightly lateral to the proteolipid ring, possibly forming part of a peripheral stalk, rather than part of the central rotor, as shown schematically in Fig. 1c.
Here we report an investigation to determine which model applies to H+-ATPases in man. We show that the human H+-ATPase d subunits are the structural orthologues of bacterial A-ATPase subunit C, despite the low level of primary sequence identity that exists between them. We also provide the first experimental evidence demonstrating that the d subunit in man forms part of the pump’s central stalk and is thus likely to be important in its rotary mechanism.
Experimental procedures
Human tissue
With ethical approval from the Cambridgeshire Local Research Ethics Committee (03/279), samples of normal human renal tissue were obtained from the discarded portions of nephrectomy specimens removed for renal tumours. Tissue was stored at −80°C until required.
Biochemicals
Taq DNA polymerase and Complete EDTA-free Protease Inhibitor Cocktail tablets were purchased from Roche Diagnostics (Burgess Hill, UK); PfuTurbo® DNA polymerase from Stratagene (La Jolla, CA, USA); restriction endonucleases, T4 DNA Ligase, pMAL-c4X and amylose resin from New England Biolabs (Hitchin, UK); pGEX-4T-1, Glutathione Sepharose 4B and ECL Plus™ from GE Healthcare (Amersham, UK); BigDye® Terminator v3.1 Cycle Sequencing reagents from Applied Biosystems (Warrington, UK); Benzonase nuclease from Novagen (Nottingham, UK); Bio-Rad Protein Assay from Bio-Rad (Hemel Hempstead, UK); horseradish peroxidase (HRP)-conjugated secondary antibodies from either Dako (Ely, UK) or Abcam (Cambridge, UK) and n-nonyl-β-d-glucopyranoside from Calbiochem (Nottingham, UK). All other chemicals were of analytical grade and purchased from either VWR (Lutterworth, UK) or Sigma-Aldrich (Gillingham, UK).
In silico structural analysis
The human H+-ATPase d1 and d2 subunits were analysed using the ensemble-based fold recognition and template modelling software Phyre (Protein Homology/analogY Recognition Engine; Bennett-Lovsey et al. 2008) available at http://www.imperial.ac.uk/phyre. This programme identifies any solved crystal structures in the Protein Data Bank (PDB) on which the protein sequence of interest can be modelled, and generates amino acid sequence alignments, secondary structure assignments and the coordinates of predicted structural models. The molecular graphics package CCP4mg (Potterton et al. 2004) was used to view the solved crystal structures identified from the PDB and the predicted structural models of d1 and d2.
Cloning of human d1, d2, D and F into bacterial expression vectors
Human kidney cDNA, prepared as described previously (Smith et al. 2005), was used as template to amplify the full-length coding regions of ATP6V0D1, ATP6V0D2, ATP6V1D and ATP6V1F with the primers listed in Table 1, using standard polymerase chain reaction (PCR) conditions. After digestion with appropriate restriction endonucleases, PCR products were ligated into either the glutathione S-transferase (GST) fusion vector pGEX-4T-1 (ATP6V0D1 and ATP6V0D2) or the maltose binding protein (MBP) fusion vector pMAL-c4X (ATP6V1D and ATP6V1F). All constructs were transformed into ultracompetent XL10-Gold Escherichia coli (Stratagene) and positive clones identified using colony PCR and restriction endonuclease digestion of plasmid DNA. Constructs that were sequence-verified using BigDye® Terminator v3.1 Cycle Sequencing were then transformed into electrocompetent BL21 E. coli provided by the Genetics Department, University of Cambridge.
Expression and purification of fusion proteins
Overnight cultures were used to inoculate 1l of either 2TY broth containing 0.1mg/ml ampicillin (GST-fusion constructs) or LB broth containing 0.2% glucose and 0.1mg/ml ampicillin (MBP-fusion constructs). Cultures were incubated at 37°C to an OD600 of 0.3–0.4 and then incubated at 30°C for 20–30min until the OD600 reached 0.6–0.8. Expression of the fusion proteins was induced by the addition of isopropyl-β-d-thiogalactopyranoside at a final concentration of 0.1mM. Following incubation at 30°C for 2h, cells were harvested at 4,000×g for 10min at 4°C and resuspended in either 50ml 1× PBS, 1mM EDTA, 1mM DTT, 1mM PMSF, 1mg/ml lysozyme, 1 Complete EDTA-free Protease Inhibitor Cocktail tablet (GST-fusion constructs) or 50ml column buffer (20mM Tris-Cl pH7.4, 200 mM NaCl, 1mM EDTA) containing 1mM PMSF, 1 mg/ml lysozyme, 1 Complete EDTA-free Protease Inhibitor Cocktail tablet (MBP-fusion constructs). Benzonase nuclease was added at 12.5U/ml and the suspensions incubated on ice for 20min, before being lysed by sonication on ice. Triton X-100 was then added at a final concentration of 1% to the GST-fusion construct lysates, with subsequent incubation on ice for 10min. All lysates were cleared of cell debris and insoluble proteins by centrifugation at 48,000×g for 30min at 4°C.
The fusion proteins were then purified by affinity chromatography as follows: lysate supernatants containing soluble proteins were incubated with either Glutathione Sepharose 4B (GST-fusion constructs) or amylose resin (MBP-fusion constructs) on a spiramixer for 2h at 4°C, washed with either 1× PBS, 300mM NaCl, 1mM DTT (GST-fusion constructs) or column buffer (MBP-fusion constructs), and eluted using either 10mM reduced glutathione, 50mM Tris-Cl pH8.0, 1mM DTT (GST-fusion constructs) or column buffer containing 10mM maltose (MBP-fusion constructs). Fractions containing fusion protein were identified by SDS-PAGE, pooled and subjected to either dialysis or buffer exchange using Vivaspin 20 (30kDa molecular weight cut off) columns (Vivascience, Epsom, UK), to remove the free glutathione or maltose. Protein concentrations were determined using the Bio-Rad Protein Assay, and SDS-PAGE was used to assess purity.
Immunoblots were carried out by standard methods to demonstrate that the fusion proteins were recognized by subunit-specific antisera. These were: rabbit anti-d1 (1:500, gift of G-H Sun-Wada, Doshisha Women’s College, Kyoto, Japan), guinea pig anti-d2 (1:1,000, SK20, Smith et al. 2005), three different goat anti-D (1:500, N-19, E-20 and Q-19, Santa Cruz Biotechnology, Heidelberg, Germany) and rabbit anti-F (1:500, FL-119, Santa Cruz Biotechnology). Membranes were blocked in PBS containing 5% nonfat dried milk and either 0.05% Tween 20 (anti-d1 and -d2) or 0.2% Tween 20 (anti-D and -F), and probed with antibodies diluted in PBS containing either 0.5% milk and 0.05% Tween 20 (anti-d1 and -d2) or 3% bovine serum albumin and 0.2% Tween 20 (anti-D and -F). Appropriate HRP-conjugated secondary antibodies were then used at a dilution of 1:7,500–15,000, before incubation with ECL Plus™ and visualization.
Pull downs from human kidney membrane using d1-GST and d2-GST
Human kidney membrane proteins (900µg), prepared as described previously (Smith et al. 2005), were incubated on a spiramixer at 4°C for 30min in solubilization buffer (10mM Tris-Cl pH7.4, 1mM EDTA, 1mM DTT, 1.5% n-nonyl-β-d-glucopyranoside, 10% glycerol and Complete EDTA-free Protease Inhibitors). Following centrifugation at 100,000×g for 1h at 4°C, the supernatant containing solubilized membrane proteins was divided into three and added to equimolar amounts of d1-GST (250µg), d2-GST (250µg) or GST (100µg), which had been immobilized on Glutathione Sepharose 4B in 1× PBS by incubation at 4°C for 2h. The samples were incubated on a spiramixer at 4°C overnight. Unbound proteins were removed by washing twice with buffer A (1× PBS, 0.1% Triton X-100), twice with buffer A containing 500mM NaCl and twice more with buffer A. Bound proteins were eluted from the sepharose beads by boiling in reducing SDS-PAGE sample buffer at 95°C for 5min. Boiled samples were centrifuged at 200×g for 1min, and the supernatants used for SDS-PAGE and immunoblot analysis.
In vitro protein interaction experiments
Equimolar amounts of D-MBP (50µg), F-MBP (40µg) or MBP (30µg) were added to equimolar amounts of d1-GST (125µg), d2-GST (125µg) or GST (50µg) immobilized on Glutathione Sepharose 4B, or to Glutathione Sepharose 4B beads alone, in buffer B (1× PBS, 0.5% Triton X-100). Samples were incubated on a spiramixer at 4°C overnight, before being washed twice with buffer B, twice with buffer B containing 500mM NaCl and twice more with buffer B to remove any unbound protein. Bound proteins were eluted from the sepharose beads as described above, and used for SDS-PAGE and immunoblot analysis.
Results
In silico structural analysis of the human H+-ATPase d1 and d2 subunits
The human H+-ATPase d1 and d2 subunits are 68% identical and share six conserved cysteine residues. They have rather low amino acid identity of 16.2% and 14.5%, respectively, with subunit C of the related A-ATPase that is present in the thermophilic bacterium T. thermophilus. They also do not share any common cysteine residues with the bacterial subunit. Despite these differences however, the eukaryotic d subunit has been proposed to be similar in structure and function to bacterial subunit C (Iwata et al. 2004). Therefore we first sought to determine whether the human d subunits are likely to be true structural orthologues of T. thermophilus subunit C.
We analysed d1 and d2 using the ensemble-based fold recognition and template modelling software Phyre (Bennett-Lovsey et al. 2008), which compares the query sequence with solved crystal structures from the PDB. This approach was used because it is generally recognised that an ensemble of multiple predictive algorithms usually has greater accuracy than any of the component individual methods (Bennett-Lovsey et al. 2008). Following submission of a query sequence, Phyre identifies any known structures in the PDB on which the query sequence can be modelled, and generates pairwise primary sequence alignments, secondary structure assignments and the coordinates of predicted structural models. The output generated by Phyre also includes an e-value and an estimated precision value. The e-value represents the number of hits that would be expected to have a match equal to or better than the reported one by chance alone. This means that the closer the e-value is to zero, the higher the significance of the match. The estimated precision value represents the percentage of times a match with a given e-value was found to be a true homologue during testing of a benchmark set of proteins and is a measure of the likelihood that the match is correct.
In our analysis, Phyre identified subunit C of T. thermophilus (monoclinic form, PDB reference: 1v9m) as being the strongest match, with e-values of 2.57 × 10−36 and 3.63× 10−37 for d1 and d2 respectively, and estimated precision values of 100%. The primary sequence alignment and secondary structure data (Fig. 2a and b) show that the predicted secondary structure of the human d subunits is remarkably conserved with that of the solved bacterial structure, despite the low level of primary sequence identity.
Primary sequence alignments and secondary structure predictions of a the human d1 subunit (Hs d1) and b the human d2 subunit (Hs d2) versus T. thermophilus subunit C (Tt C). The match line for the primary sequence alignment was generated using the Perl module Bio::AlignIO and is defined as per the same amino acid conservation and substitution criterion set for ClustalW. It shows the position of identical residues (*); conserved substitutions (:) and semi-conserved substitutions (.). A three-state secondary structure assignment is displayed on each sequence and indicates the position of alpha-helices (magenta), beta-sheets (brown; d2 residues 7–10) and random coils (grey). The relative positions of the three different domains of T. thermophilus subunit C are indicated on the match line as follows: domain 1 (blue), domain 2 (green) and domain 3 (plum). N- and C-termini are both located in domain 1
The programme CCP4mg (Potterton et al. 2004) was used to view and compare the predicted structural models of d1 (Fig. 3a) and d2 (Fig. 3c) and the solved structure of the bacterial subunit C (Fig. 3e). The three proteins have an almost identical alpha-helical three-domain structure, with the N- and C-termini both being located in domain 1. Residues 179–185 in the bacterial sequence were found to be disordered in the crystal structure (Numoto et al. 2004), and are missing from the model as indicated by the arrow (Fig. 3e); residues 1–4 were also disordered. The predicted structures of d1 and d2 were then superimposed on subunit C (shown in grey) to highlight potential structural differences (Fig. 3b and d). This reveals very few likely differences, most of which affect regions made up of random coils. The remarkable level of predicted structural conservation that exists between these proteins suggests they are indeed structural orthologues.
Ribbon models of the predicted structures of the human d1 (a) and d2 (c) subunits, and the solved structure of T. thermophilus subunit C (e), are shown viewed from the side. Domains 1, 2 and 3 are coloured blue, green and plum, respectively. The positions of N- and C-termini are indicated. Residues 179–185 in the bacterial orthologue, located at the boundary of domains 2 and 3, were disordered in the crystal structure (Numoto et al. 2004) and are missing from the model, as indicated by the arrow (e). Residues 1–4 were also disordered. To highlight potential structural differences, the predicted structures of d1 (b) and d2 (d) were superimposed on the subunit C structure, which is shown in grey
Expression and purification of d1-GST and d2-GST
We wanted to determine whether the human d subunits are able to interact with the H+-ATPase central stalk subunits D and F, which would be expected if the d subunit has a similar role to that of the T. thermophilus subunit C, as well as being its structural counterpart. Thus, full-length human d1 and d2 were cloned into the GST-fusion protein vector pGEX-4T-1, successfully expressed in BL21 E. coli and purified using affinity chromatography. SDS-PAGE analysis showed that d1-GST and d2-GST both run as a single band with an apparent molecular weight of approximately 55kDa (Fig. 4a). Although this is lower than the predicted molecular weight of around 68kDa, similar aberrant migration of the yeast d subunit in SDS-PAGE has been reported (Bauerle et al. 1993). Immunoblotting demonstrated that d1-GST and d2-GST are recognized by subunitspecific antisera (Fig. 4b).
d1 and d2 pull downs using human kidney membrane. a SDSPAGE analysis of affinity purified d1-GST and d2-GST shows that they both run as a single band at approximately 55 kDa. b Immunoblotting shows that they are recognized by anti-d1- and antid2-specific antisera, respectively. c Immunoblotting using specific anti-D (N-19) and anti-F antisera shows that d1-GST or d2-GST, but not GST alone are able to pull down the D and F subunits from human kidney membrane preparations (indicated by the arrowheads). Human kidney membrane acts as a positive control. Molecular weight markers are in kilodalton
d1 and d2 pull down the H+-ATPase D and F subunits from human kidney membrane
We then immobilized equimolar amounts of each of the affinity purified fusion proteins and GST alone on Glutathione Sepharose 4B, and demonstrated that d1-GST and d2-GST, but not GST alone, are each able to pull down the H+-ATPase D and F subunits from solubilized human kidney membrane preparations (Fig. 4c). This shows that d1 and d2 can interact with the proton pump complex, but by itself does not necessarily provide evidence of a direct interaction with the D and F subunits.
d1 and d2 interact directly with the D and F subunits
We therefore next sought to determine whether the human D and F subunits are able to interact directly with d1 and d2. Full length human D and F were cloned into the MBP-fusion protein vector pMAL-c4X and successfully expressed in BL21 E. coli. Affinity purified D-MBP and F-MBP were shown by SDS-PAGE to have apparent molecular weights of approximately 70 and 50kDa, respectively (Fig. 5a).
Expression and purification of D-MBP and F-MBP. a SDS-PAGE analysis of affinity purified D-MBP and F-MBP shows that each runs as a single major species, at approximately 70 and 50 kDa, respectively. b In immunoblotting, D-MBP was detected using two different anti-D-specific polyclonal antisera, E-20 recognizing an internal epitope and Q-19 recognizing a C terminal epitope. The weak lower band corresponding to D-MBP seen in the left lane of the SDSPAGE gel (a) was recognized by E-20, but not by Q-19, suggesting that a small proportion of D-MBP is missing part of the C terminus. c F-MBP was recognized by an F subunit-specific antiserum. Molecular weight markers are in kilodalton
D-MBP was immunoblotted using two different anti-D subunit-specific polyclonal antisera, one that recognizes an internal epitope (E-20) and one that recognizes a C terminal epitope (Q-19), as shown in Fig. 5b. The weak lower band corresponding to D-MBP seen in the left lane of the SDS-PAGE gel (Fig. 5a) was recognized by E-20, but not by Q-19, suggesting that it is missing part of the C terminus. F-MBP was recognized by an F subunit-specific antiserum (Fig. 5c).
We then carried out in vitro protein interaction experiments, wherein equimolar amounts of d1-GST, d2-GST or GST alone, immobilized on Glutathione Sepharose 4B, were incubated with equimolar amounts of either D-MBP, F-MBP or MBP alone. Immunoblots using anti-D and anti-F antisera show that d1-GST and d2-GST, but not GST or beads alone, are able directly to pull down D-MBP (Fig. 6a) and F-MBP (Fig. 6b). Control immunoblots using an anti-MBP antiserum demonstrate that this interaction is specific (Fig. 6c and d) and not due to the binding of MBP alone by d1-GST and d2-GST. The doublet corresponding to D-MBP seen in Fig. 6c results from the fact that the anti-MBP antibody can recognize both full length D-MBP and the slightly truncated form mentioned above. These data show that human d1 and d2 are able to directly interact with the D and F subunits.
In vitro interaction experiments. Immunoblots using subunit-specific antisera show that d1-GST (lane 1) and d2-GST (lane 2), but not GST (lane 3) or Glutathione Sepharose 4B beads alone (lane 4), can directly pull down D-MBP (a) and F-MBP (b) as indicated by the arrowheads. Control immunoblots using an anti-MBP antiserum were carried out for d1-GST (lanes 1, 5), d2-GST (lanes 2, 6), GST (lanes 3, 7) and beads only (lanes 4, 8) incubated with D-MBP/MBP (c) or with F-MBP/MBP (d), as indicated. These show that the interaction between the subunits is specific and does not solely result from binding of MBP by d1-GST and d2-GST. The doublet seen in lanes 1 and 2 in c is due to the anti-MBP antibody recognizing both full length and the slightly truncated form of D-MBP. Molecular weight markers are in kilodalton
Discussion
The vacuolar H+-ATPase d subunit is known to associate with the integral membrane V0 domain (Bauerle et al. 1993), but its function and precise position within the pump complex are still under debate. Two hypotheses regarding its localization have been put forward that predicate potentially different roles, the first suggests that it is similar to bacterial A-ATPase subunit C and constitutes part of the central rotor (Iwata et al. 2004), and the second, based on yeast studies, proposes a more lateral position as part of a peripheral stalk (Thaker et al. 2007). The data from our study indicate that the human d1 and d2 subunits are centrally located within the pump, thereby supporting the first hypothesis.
Using in silico structural analysis, we have here demonstrated that the human H+-ATPase d subunits show remarkable structural conservation with T. thermophilus A-ATPase subunit C, despite their low level of primary sequence identity, indicating that these proteins are likely to be true structural, and presumably functional, orthologues. Therefore, despite the extensive sequence divergence that has occurred, these subunits must have been subjected to strong structural and functional evolutionary pressure during the evolution of bacteria and man.
The solved structure of bacterial subunit C indicates that the protein has an alpha-helical three-domain structure and an overall shape resembling a bottle-stopper. Cross-linking experiments have shown that it is in close proximity to the L subunits of the proteolipid ring, which correspond to the c, c′ and c″ subunits of the eukaryotic H+-ATPase, as well as subunit F of the central stalk (Iwata et al. 2004). Further cross-linking experiments have shown that subunits D and F are in turn in close proximity to each other (Makyio et al. 2005). It has thus been proposed that subunit C has a socket-like function attaching the central stalk subunits D and F to the proteolipid ring. If the human d subunits have a similar role to bacterial subunit C, one would predict that they should interact with D and F, and also with components of the proteolipid ring. In this study we have provided the first experimental evidence showing that d1 and d2 do indeed interact directly with the D and F subunits of the human H+-ATPase. Unfortunately we are presently unable to determine whether the human d subunits are also able to bind to the proteolipid subunits, since there are no antibodies available that recognize these proteins, nor have these proteins ever been successfully expressed in vitro.
Our data do not support the alternative hypothesis—that the eukaryotic d subunit is located above and lateral to the proteolipid ring, without any connection to it, possibly forming part of a peripheral stalk, as suggested by Thaker et al. (2007). These authors observed structural differences between the bacterial subunit C and the yeast d subunit, with the latter being not only larger, but also more anisometric than subunit C. However, the apparent differences might be due to methodological variation, as they were comparing the subunit C crystal structure with a low-resolution solution structure of the yeast d subunit etermined from small-angle X-ray scattering data. They also suggest that the inability of the T. thermophilus orthologue to rescue the phenotype of the yeast d subunit deletion strain (Owegi et al. 2006) indicates that the proteins are not functional equivalents. This is not necessarily the case as there are several examples of eukaryotic H+-ATPase subunits being unable functionally to complement the orthologous yeast subunit deletion strain, for example the mouse G1 and G3 subunits (Sun-Wada et al. 2003) and the human a4 subunit (Su et al. 2008).
The idea of peripheral stalk residency for the d subunit came in part from consideration of the 3D structural model of the bovine H+-ATPase proton channel generated by Wilkens and Forgac (2001), in which there are two protein densities covering the cytoplasmic opening of the channel. Although as described earlier, the more peripheral one is that proposed by Thaker and collaborators to correspond to subunit d (Fig. 1), Wilkens and collaborators suggested that the central density has a clear three-domain structure like bacterial subunit C (Fig. 7) and is thus likely to be the d subunit (Wilkens et al. 2005). Our data support this model for localization of the d subunit within the pump complex. If it forms part of the central stalk, it is likely to play a role in the rotary mechanism of the H+-ATPase, and therefore be important in the coupling of ATP hydrolysis and proton transport as has been proposed recently (Nishi et al. 2003; Owegi et al. 2006).
The ribbon model of T. thermophilus subunit C, viewed from the top, shows the protein’s alpha-helical three-domain structure. Looking at the 3D structural model of the bovine H+-ATPase V0 domain from the top (Wilkens and Forgac 2001), it can be seen that the central density, covering the cytoplasmic opening of the proton channel, has a similar three-domain structure (indicated by asterisks), suggesting that it corresponds to subunit C and therefore the eukaryotic d subunit
It has been suggested that in yeast, the formation of a disulphide bond between Cys36 and Cys329 is important for folding (Thaker et al. 2007). These cysteines are conserved across all eukaryotic species including man, but not in T. thermophilus. However, how important disulphide bond formation is to the function of subunit d is unclear, since the protein is normally present in the reducing environment of the cytoplasm where such bonds would not usually persist. Indeed, this bond had little effect on in vitro tertiary structure. Also, a yeast strain expressing a C329A d subunit mutation retained 72% proton transport and 41% ATPase activity levels relative to wild type at its vacuolar membrane (Owegi et al. 2006), suggesting that the substitution is not highly disruptive. Furthermore, the bond between Cys267 and Cys322 seen in both hexagonal and monoclinic forms of the bacterial crystal structure (Iwata et al. 2004; Numoto et al. 2004), was proposed by Iwata to be an artefact formed during sample preparation under aerobic conditions. In addition, bacterial H+-ATPases containing mutant subunit C with both cysteines replaced by serine are properly assembled and fully functional (Iwata et al. 2004). All these data suggest that disulphide bond formation is not critical to the function of these orthologous proteins in their native cellular environments.
In summary, our data indicate that the d subunit in man forms part of the central rotor of the H+-ATPase. The determination of the location of proton pump components is important, since in order to facilitate the therapeutic targeting of the H+-ATPase in diseases such as osteoporosis and cancer, it is essential to understand the proper isoform composition and precise localization of subunits within the V1/V0 complex. Such information may aid the development of small molecules that can target exposed regions of the complex in an isoform-specific manner. For example, the possible exclusive co-expression of the a3 and d2 subunit isoforms in the ruffled border of osteoclasts, described previously, (Smith et al. 2005), may in the future allow the design of antiresorptive agents that can specifically target this specialized proton pump in the treatment of osteoporosis.
References
Bauerle C, Ho MN, Lindorfer MA, Stevens TH (1993) J Biol Chem 268:12749–12757
Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA (2008) Proteins 70:611–625
Blair HC, Teitelbaum SL, Ghiselli R, Gluck S (1989) Science 245:855–857
Forgac M (2007) Nat Rev Mol Cell Biol 8:917–929
Hirata T, Iwamoto-Kihara A, Sun-Wada GH, Okajima T, Wada Y, Futai M (2003) J Biol Chem 278:23714–23719
Imamura H, Nakano M, Noji H, Muneyuki E, Ohkuma S, Yoshida M, Yokoyama K (2003) Proc Natl Acad Sci U S A 100:2312–2315
Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, Makyio H, Hankamer B, Barber J, Yoshida M, Yokoyama K, Iwata S (2004) Proc Natl Acad Sci U S A 101:59–64
Makyio H, Iino R, Ikeda C, Imamura H, Tamakoshi M, Iwata M, Stock D, Bernal RA, Carpenter EP, Yoshida M, Yokoyama K, Iwata S (2005) EMBO J 24:3974–3983
Nishi T, Kawasaki-Nishi S, Forgac M (2003) J Biol Chem 278:46396–46402
Numoto N, Kita A, Miki K (2004) Acta Crystallogr D Biol Crystallogr 60:810–815
Owegi MA, Pappas DL, Finch MW Jr, Bilbo SA, Resendiz CA, Jacquemin LJ, Warrier A, Trombley JD, McCulloch KM, Margalef KL, Mertz MJ, Storms JM, Damin CA, Parra KJ (2006) J Biol Chem 281:30001–30014
Pastor-Soler N, Pietrement C, Breton S (2005) Physiology (Bethesda) 20:417–428
Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M (2004) Acta Crystallogr D Biol Crystallogr 60:2288–2294
Sennoune SR, Martinez-Zaguilan R (2007) J Bioenerg Biomembr 39:427–433
Smith AN, Borthwick KJ, Karet FE (2002) Gene 297:169–177
Smith AN, Jouret F, Bord S, Borthwick KJ, Al-Lamki RS, Wagner CA, Ireland DC, Cormier-Daire V, Frattini A, Villa A, Kornak U, Devuyst O, Karet FE (2005) J Am Soc Nephrol 16:1245–1256
Su Y, Blake-Palmer KG, Sorrell S, Javid B, Bowers K, Zhou A, Chang SH, Qamar S, Karet FE (2008) Am J Physiol Renal Physiol. doi:10.1152/ajprenal.90258.2008
Sun-Wada GH, Yoshimizu T, Imai-Senga Y, Wada Y, Futai M (2003) Gene 302:147–153
Thaker YR, Roessle M, Gruber G (2007) J Bioenerg Biomembr 39:275–289
Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP (2004) Physiol Rev 84:1263–1314
Wilkens S, Forgac M (2001) J Biol Chem 276:44064–44068
Wilkens S, Zhang Z, Zheng Y (2005) Micron 36:109–126
Acknowledgements
We are grateful to Prof. S. Wilkens for kindly providing the 3D reconstruction images of the V0 domain. This work was supported by the Wellcome Trust, of which FEK is a Senior Clinical Research Fellow. CIMR is in receipt of a Wellcome Trust Strategic Award (079895).
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Smith, A.N., Francis, R.W., Sorrell, S.L. et al. The d subunit plays a central role in human vacuolar H+-ATPases. J Bioenerg Biomembr 40, 371–380 (2008). https://doi.org/10.1007/s10863-008-9161-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10863-008-9161-y