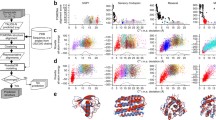
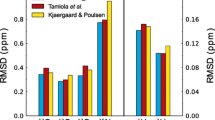
Abstract
Protein structure determination using nuclear magnetic resonance (NMR) spectroscopy can be both time-consuming and labor intensive. Here we demonstrate how chemical shift threading can permit rapid, robust, and accurate protein structure determination using only chemical shift data. Threading is a relatively old bioinformatics technique that uses a combination of sequence information and predicted (or experimentally acquired) low-resolution structural data to generate high-resolution 3D protein structures. The key motivations behind using NMR chemical shifts for protein threading lie in the fact that they are easy to measure, they are available prior to 3D structure determination, and they contain vital structural information. The method we have developed uses not only sequence and chemical shift similarity but also chemical shift-derived secondary structure, shift-derived super-secondary structure, and shift-derived accessible surface area to generate a high quality protein structure regardless of the sequence similarity (or lack thereof) to a known structure already in the PDB. The method (called E-Thrifty) was found to be very fast (often < 10 min/structure) and to significantly outperform other shift-based or threading-based structure determination methods (in terms of top template model accuracy)—with an average TM-score performance of 0.68 (vs. 0.50–0.62 for other methods). Coupled with recent developments in chemical shift refinement, these results suggest that protein structure determination, using only NMR chemical shifts, is becoming increasingly practical and reliable. E-Thrifty is available as a web server at http://ethrifty.ca.




Similar content being viewed by others
References
Alexander PA, He Y, Chen Y, Orban J, Bryan PN (2009) A minimal sequence code for switching protein structure and function. Proc Natl Acad Sci 106(50):21149–21154
Altschul SF, Gish W (1996) Local alignment statistics. Meth Enzymol 266:460–480
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Berjanskii MV, Wishart DS (2005) A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc 127(43):14970–14971
Berjanskii MV, Wishart DS (2013) A simple method to measure protein side-chain mobility using NMR chemical shifts. J Am Chem Soc 135(39):14536–14539
Berjanskii MV, Neal S, Wishart DS (2006) PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res 34(suppl 2):W63-W69
Berjanskii M, Tang P, Liang J, Cruz JA, Zhou J, Zhou Y et al (2009) GeNMR: a web server for rapid NMR-based protein structure determination. Nucleic Acids Res 37(suppl 2):W670-W677
Berjanskii M, Arndt D, Liang Y, Wishart DS (2015) A robust algorithm for optimizing protein structures with NMR chemical shifts. J Bio NMR 63(3):255–264
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Betancourt MR, Skolnick J (2001) Universal similarity measure for comparing protein structures. Biopolymers 59(5):305–309
Bonomi M, Heller GT, Camilloni C, Vendruscolo M (2017) Principles of protein structural ensemble determination. Curr Opin Struct Biol 42:106–116
Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL (2012) Domain enhanced lookup time accelerated BLAST. Biol Direct 7(1):12
Bowie JU, Luthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164–170
Camilloni C, Vendruscolo M (2014) Statistical mechanics of the denatured state of a protein using replica-averaged metadynamics. J Am Chem Soc 136(25):8982–8991
Cavalli A, Salvatella X, Dobson CM, Vendruscolo M (2007) Protein structure determination from NMR chemical shifts. Proc Natl Acad Sci 104(23):9615–9620
Choy WY, Forman-Kay JD (2001) Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J Mol Biol 308:1011–1032
Eddy SR (1998) Profile hidden markov models. Bioinformatics 14:755–763
Ginzinger SW, Fischer J (2006) SimShift: identifying structural similarities from NMR chemical shifts. Bioinformatics 22(4):460–465
Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M et al (2007) The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res 35:D291–D297
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273(1):283–298
Gutmanas A, Adams PD, Bardiaux B, Berman HM, Case DA, Fogh RH et al (2015) NMR exchange format: a unified and open standard for representation of NMR restraint data. Nat Struct Mol Biol 22(6):433–434
Hafsa NE, Wishart DS (2014) CSI 2.0: a significantly improved version of the Chemical Shift Index. J Biomol NMR 60(2–3):131–146
Hafsa NE, Arndt D, Wishart DS (2015a) CSI 3.0: a web server for identifying secondary and super-secondary structure in proteins using NMR chemical shifts. Nucleic Acids Res 43(W1):W370–W377
Hafsa NE, Arndt D, Wishart DS (2015b) Accessible surface area from NMR chemical shifts. J Biomol NMR 62(3):387–401
Han B, Liu Y, Ginzinger SW, Wishart DS (2011) SHIFTX2: significantly improved protein chemical shift prediction. J Biomol NMR 50(1):43–57
Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci 89(22):10915–10919
Holm L, Rosenström P (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38(suppl 2):W545-W549
Jones DT, Taylor WR, Thornton JM (1992) A new approach to protein fold recognition. Nature 358:86–89
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22(12):2577–2637
Karplus K, Barrett C, Hughey R (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics 14(10):846–856
Krogh A, Brown M, Mian IS, Sjölander K, Haussler D (1994) Hidden markov models in computational biology—applications to protein modeling. J Mol Biol 235:1501–1531
Kuszewsk J, Gronenborn AM, Clore GM (1995) The impact of direct refinement against proton chemical shifts on protein structure determination by NMR. J Magn Reson 107:293–297
Lindahl E, Elofsson A (2000) Identification of related proteins on family, superfamily and fold level. J Mol Biol 295(3):613–625
Madhusudhan MS, Marti-Renom MA, Sanchez R, Sali A (2006) Variable gap penalty for protein sequence–structure alignment. Protein Eng Des Sel 19(3):129–133
Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res 32(2):W590–W594
Medek A, Hajduk PJ, Mack J, Fesik SW (2000) The use of differential chemical shifts for determining the binding site location and orientation of protein-bound ligands. J Am Chem Soc 122(6):1241–1242
Mielke SP, Krishnan VV (2009) Characterization of protein secondary structure from NMR chemical shifts. Prog Nucl Magn Reson Spectrosc 54(3):141–165
Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247(4):536–540
Ortiz AR, Strauss CE, Olmea O (2002) MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci 11(11):2606–2621
Osapay K, Case DA (1991) A new analysis of proton chemical shifts in proteins. J Am Chem Soc 113(25):9436–9444
Osapay K, Theriault Y, Wright PE, Case DA (1994) Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints. J Mol Biol 244:183–197
Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, DeBolt S et al (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91(1):1–41
Peng J, Xu J (2010) Low-homology protein threading. Bioinformatics 26(12):i294-i300
Rosato A, Vranken W, Fogh RH, Ragan TJ, Tejero R, Pederson K, Vuister GW (2015) The second round of critical assessment of automated structure determination of proteins by NMR: CASD-NMR-2013. J Biomol NMR 62(4):413–424
Rost B (1995) TOPITS: threading one-dimensional predictions into three-dimensional structures. Proc Int Conf Intell Syst Mol Biol 3:314–321
Rost B, Schneider R, Sander C (1997) Protein fold recognition by prediction-based threading. J Mol Biol 270(3):471–480
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ (2001) Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123(13):2970–2978
Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160(1):65–73
Sharma D, Rajarathnam K (2000) C-13 NMR chemical shifts can predict disulfide bond formation. J Biomol NMR 18:165–171
Shen Y, Bax A (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR 56(3):227–241
Shen Y, Bax A (2015) Homology modeling of larger proteins guided by chemical shifts. Nat Methods 12(8):747–750
Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G et al (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc Natl Acad Sci 105(12):4685–4690
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44(4):213–223
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7(1):539
Siew N, Elofsson A, Rychlewski L, Fischer D (2000) MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics 16(9):776–785
Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Mol Biol 147(1):195–197
Söding J (2005) Protein homology detection by HMM–HMM comparison. Bioinformatics 21(7):951–960
Song Y, DiMaio F, Wang RYR, Kim D, Miles C, Brunette TJ et al (2013) High-resolution comparative modeling with RosettaCM. Structure 21(10):1735–1742
Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J et al (2008) BioMagResBank. Nucleic Acids Res 36(suppl 1):D402–D408
Vranken WF, Rieping W (2009) Relationship between chemical shift value and accessible surface area for all amino acid atoms. BMC Struct Biol 9(1):20
Wang G, Dunbrack RL (2003) PISCES: a protein sequence culling server. Bioinformatics 19(12):1589–1591
Wishart DS (2011) Interpreting protein chemical shift data. Prog Nucl Magn Reson Spectrosc 58(1):62–87
Wishart DS, Case DA (2001) Use of chemical shifts in macromolecular structure determination. Methods Enzymol 338:3–34
Wishart DS, Nip AM (1998) Protein chemical shift analysis: a practical guide. Biochem Cell Biol 76:153–163
Wishart DS, Sykes BD (1994a) Chemical shifts as a tool for structure determination. Methods Enzymol 239:363–392
Wishart DS, Sykes BD (1994b) The 13C chemical shift index: a simple method for the identification of protein secondary structure using 13C chemical shift data. J Biomol NMR 4(2):171–180
Wishart DS, Sykes BD, Richards FM (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 31(6):1647–1651
Wishart DS, Arndt D, Berjanskii M, Tang P, Zhou J, Lin G (2008) CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res 36:W496–W502
Xu J, Zhang Y (2010) How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26(7):889–895
Zhang Y, Skolnick J (2004) Scoring function for automated assessment of protein structure template quality. Proteins 57:702–710
Acknowledgements
Financial support from the Natural Sciences and Engineering Research Council (NSERC), the Alberta Prion Research Institute (APRI) and the Canadian Institutes of Health Research (CIHR) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hafsa, N.E., Berjanskii, M.V., Arndt, D. et al. Rapid and reliable protein structure determination via chemical shift threading. J Biomol NMR 70, 33–51 (2018). https://doi.org/10.1007/s10858-017-0154-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-017-0154-1