Abstract
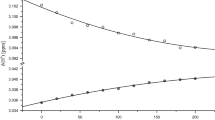
For a better understanding of nuclear magnetic resonance (NMR) detected pressure responses of folded as well as unstructured proteins the availability of data from well-defined model systems are indispensable. In this work we report the pressure dependence of chemical shifts of the backbone atoms 1Hα, 13Cα and 13C′ in the protected tetrapeptides Ac-Gly-Gly-Xxx-Ala-NH2 (Xxx one of the 20 canonical amino acids). Contrary to expectation the chemical shifts of these nuclei have a nonlinear dependence on pressure in the range from 0.1 to 200 MPa. The polynomial pressure coefficients B 1 and B 2 are dependent on the type of amino acid studied. The coefficients of a given nucleus show significant linear correlations suggesting that the NMR observable pressure effects in the different amino acids have at least partly the same physical cause. In line with this observation the magnitude of the second order coefficients of nuclei being direct neighbors in the chemical structure are also weakly correlated.
Graphical Abstract






Similar content being viewed by others
References
Akasaka K (2006) Probing conformational fluctuation of proteins by pressure perturbation. Chem Rev 106:1814–1835
Akasaka K, Li H, Yamada H, Li R, Thoresen T, Woodward CK (1999) Pressure response of protein backbone structure. Pressure-induced amide 15N chemical shifts in BPTI. Protein Sci 8:1946–1953
Arnold MR, Kremer W, Lüdemann HD, Kalbitzer HR (2002) 1H-NMR parameters of common amino acid residues measured in aqueous solutions of the linear tetrapeptides Gly-Gly-X-Ala at pressures between 0.1 and 200 MPa. Biophys Chem 96:129–140
Asakawa N, Kameda T, Kuroki S, Kurosu H, Ando S, Ando I, Shoji A (1998) Structural studies of hydrogen-bonded peptides and polypeptides by solid state NMR. Ann Rep NMR Spectrosc 35:55–137
Avbelj F, Kocjan D, Baldwin RL (2004) Protein chemical shifts arising from α-helices and β-sheets depend on solvent exposure. Proc Natl Acad Sci 50:17394–17397
Braun D, Wider G, Wüthrich K (1994) Sequence-corrected 15N random coil chemical shifts. J Am Chem Soc 116:8466–8469
Buckingham AD (1960) Chemical shifts in the nuclear magnetic resonance spectra of molecules containing polar groups. Can J Chem 38:300–307
Bundi A, Wüthrich K (1979) 1H-NMR parameters of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers 18:285–297
Cordier F, Barfield M, Stephan Grzesiek S (2003) Direct Observation of Cα–Hα···O=C hydrogen bonds in proteins by interresidue h3JCαC′ scalar couplings. J Am Chem Soc 125:15750–15751
De Simone A, Cavalli A, Hsu STD, Vranken W, Vendruscolo M (2009) Accurate random coil chemical shifts from an analysis of loop regions in native states of proteins. J Am Chem Soc 131:16332–16333
Erlach MB, Munte CE, Kremer W, Hartl R, Rochelt D, Niesner D, Kalbitzer HR (2010) Ceramic cells for high pressure NMR spectroscopy of proteins. J Magn Reson 204:196–199
Erlach MB, Koehler J, Moeser B, Horinek D, Kremer W, Kalbitzer HR (2014) Relationship between nonlinear pressure-induced chemical shift changes and thermodynamic parameters. J Phys Chem B 118:5681–5690
Frach R, Kibies P, Böttcher S, Pongratz T, Strohfeldt S, Kurrmann S, Koehler J, Hofmann M, Kremer W, Kalbitzer HR, Reiser O, Horinek D, Kast SM (2016) The Chemical Shift Baseline for High-Pressure NMR Spectroscopy of Proteins. Angew Chemie. doi:10.1002/anie.201602054
Gronwald W, Kalbitzer HR (2004) Automated structure determination of proteins by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 44:33–96
Gross M, Jaenicke R (1994) Proteins under pressure. The influence of high hydrostatic pressure on structure, function and assembly of proteins and protein complexes. Eur J Biochem 221:617–630
Groß KH, Kalbitzer HR (1988) Distribution of chemical shifts in 1H nuclear magnetic resonance spectra of proteins. J Magn Reson 76:87–99
Jimenez MA, Nieto JL, Rico M, Santoro J, Herranz J, Bermejo FJ (1986) A study of the NH NMR signals of Gly-Gly-X-Ala tetrapeptides in H2O at low temperature. J Mol Struct 143:435–438
Kachel N, Kremer W, Zahn R, Kalbitzer HR (2006) Observation of intermediate states of the human prion protein by high pressure NMR spectroscopy. BMC Struct Biol 16:6
Kalbitzer HR (2015) High pressure NMR methods for characterizing functional substates of proteins. In: Akasaka K, Matsuki H (eds) High pressure bioscience—basic concepts, applications and frontiers. Springer, Heidelberg, pp 179–198
Kalbitzer HR, Görler A, Li H, Dubovskii P, Hengstenberg W, Kowolik C, Yamada H, Akasaka K (2000) 15N and 1H NMR study of histidine containing protein (HPr) from Staphylococcus carnosus at high pressure. Protein Sci 9:693–703
Kalbitzer HR, Spoerner M, Ganser P, Hosza C, Kremer W (2009) Fundamental link between folding states and functional states of proteins. J Am Chem Soc 131:16714–16719
Kalbitzer HR, Rosnizeck IC, Munte CE, Puthenpurackal Narayanan S, Kropf V, Spoerner M (2013) Intrinsic allosteric inhibition of signaling proteins by targeting rare interaction states detected by high-pressure NMR spectroscopy. Angew Chem Int Ed 52:14242–14246
Kitahara R, Hata K, Li H, Williamson MP, Akasaka K (2013) Pressure-induced chemical shifts as probes for conformational fluctuations in proteins. Prog Nucl Magn Reson Spectrosc 71:35–58
Kjaergaard M, Poulsen FM (2011) sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J Biomol NMR 50:157–165
Kjaergaard M, Brander S, Poulsen FM (2011) Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J Biomol NMR 49:139–149
Koehler J, Erlach MB, Crusca E, Kremer W, Munte CE, Kalbitzer HR (2012) Pressure dependence of 15N chemical shifts in model peptides Ac-Gly-Gly-X-Ala-NH2. Materials 5:1774–1786
Kurrmann S (2015) Hochdruck-NMR-Spektroskopie an Thrimethylamin-N-Oxid und N-Methyl-Acetamid. Bachelor thesis, University of Regensburg
Le HB, Oldfield E (1994) Correlation between 15N NMR chemical-shifts in proteins and secondary structure. J Biomol NMR 4:341–348
Li H, Yamada H, Akasaka K (1998) Effect of pressure on individual hydrogen bonds in proteins. Basic pancreatic trypsin inhibitor. Biochemistry 37:1167–1173
Li H, Yamada H, Akasaka K, Gronenborn AM (2000) Pressure alters electronic orbital overlap in hydrogen bonds. J Biomol NMR 18:207–216
Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. Pure Appl Chem 70:117–142
Merutka G, Dyson HJ, Wright PE (1995) ‘Random coil’ 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J Biomol NMR 5:14–24
Peterson RW, Wand AJ (2005) Self-contained high pressure cell, apparatus, and procedure for the preparation of encapsulated proteins dissolved in low viscosity fluids for nuclear magnetic resonance spectroscopy. Rev Sci Instrum 76:094101
Peti W, Smith LJ, Redfield C, Schwalbe H (2001) Chemical shifts in denatured proteins: resonance assignments for denatured ubiquitin and comparisons with other denatured proteins. J Biomol NMR 19:153–165
Platzer G, Okon M, McIntosh LP (2014) pH-dependent random coil 1H, 13C, and 15N chemical shifts of the ionizable amino acids: a guide for protein pK a measurements. J Biomol NMR 60:109–129
Plaxco KW, Morton CJ, Grimshaw SB, Jones JA, Pitkeathly M, Campbell ID, Dobson CM (1997) The effects of guanidine hydrochloride on the ‘random coil’ conformations and NMR chemical shifts of the peptide series GGXGG. J Biomol NMR 10:221–230
R Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Raiford DS, Fisk CL, Becker ED (1979) Calibration of methanol and ethylene glycol nuclear magnetic resonance thermometers. Anal Chem 51:2050–2051
Richarz R, Wüthrich K (1978) Carbon-13 NMR chemical shifts of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers 17:2133–2141
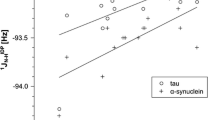
Roche J, Ying J, Maltsev AS, Bax A (2013) Impact of hydrostatic pressure on an intrinsically disordered protein: a high-pressure NMR study of a-synuclein. ChemBioChem 14:1754–1761
Rosnizeck IC, Graf T, Spoerner M, Tränkle J, Filchtinski D, Herrmann C, Gremer L, Vetter IR, Wittinghofer A, König B, Kalbitzer HR (2010) Stabilizing a weak binding state for effectors in the human Ras-protein by cyclen complexes. Angew Chem Int Ed 49:3830–3833
Rosnizeck IC, Spoerner M, Harsch T, Kreitner S, Filchtinski D, Herrmann C, Engel D, König B, Kalbitzer HR (2012) Metal–bis(2-picolyl)amine complexes as state 1(T) inhibitors of activated Ras protein. Angew Chem Int Ed 51:10647–10651
Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ (2000) Random coil chemical shifts in acidic 8M urea: implementation of random coil shift data in NMRView. J Biomol NMR 18:43–48
Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ (2001) Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123:2970–2978
Tamiola K, Acar B, Mulder FAA (2010) Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc 132:18000–18003
Thanabal V, Omecinsky DO, Reily MD, Cody WL (1994) The 13C chemical shifts of amino acids in aqueous solution containing organic solvents: application to the secondary structure characterization of peptides in aqueous trifluoroethanol solution. J Biomol NMR 4:47–59
Wagner G, Pardi A, Wüthrich K (1983) Protein conformation and proton nuclear magnetic-resonance chemical shifts. J Am Chem Soc 105:5948
Wang Y, Jardetzky O (2002) Probability-based protein secondary structure identification using combined NMR chemical-shift data. Protein Sci 11:852–861
Wishart DS, Sykes BD (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. J Biomol NMR 31:1647–1651
Wishart DS, Sykes BD (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:171–180
Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD (1995a) 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR 5:67–82
Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD (1995b) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140
Zhang H, Neal S, Wishart DS (2003) RefDB: a database of uniformly referenced protein chemical shifts. J Biomol NMR 25:173–195
Acknowledgments
This work has been supported by the DFG (FOR1979 and KA 647), the Humboldt Society, Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2010/01362-5 and 2010/12953-4), and the Human Frontier Science Program Organization (HFSPO).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Erlach, M.B., Koehler, J., Crusca, E. et al. Pressure dependence of backbone chemical shifts in the model peptides Ac-Gly-Gly-Xxx-Ala-NH2 . J Biomol NMR 65, 65–77 (2016). https://doi.org/10.1007/s10858-016-0030-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-016-0030-4