Abstract
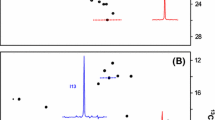
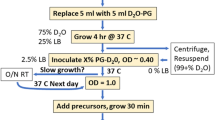
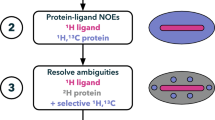
The specific protonation of valine and leucine methyl groups in proteins is typically achieved by overexpressing proteins in M9/D2O medium supplemented with either labeled α-ketoisovalerate for the labeling of the four prochiral methyl groups or with 2-acetolactate for the stereospecific labeling of the valine and leucine side chains. However, when these labeling schemes are applied to large protein assemblies, significant overlap between the correlations of the valine and leucine methyl groups occurs, hampering the analysis of 2D methyl-TROSY spectra. Analysis of the leucine and valine biosynthesis pathways revealed that the incorporation of labeled precursors in the leucine pathway can be inhibited by the addition of exogenous l-leucine-d10. We exploited this property to label stereospecifically the pro-R and pro-S methyl groups of valine with minimal scrambling to the leucine residues. This new labeling protocol was applied to the 468 kDa homododecameric peptidase TET2 to decrease the complexity of its NMR spectra. All of the pro-S valine methyl resonances of TET2 were assigned by combining mutagenesis with this innovative labeling approach. The assignments were transferred to the pro-R groups using an optimally labeled sample and a set of triple resonance experiments. This improved labeling scheme enables us to overcome the main limitation of overcrowding in the NMR spectra of prochiral methyl groups, which is a prerequisite for the site-specific measurement of the structural and dynamic parameters or for the study of interactions in very large protein assemblies.






Similar content being viewed by others
References
Amero C, Schanda P, Durá MA, Ayala I, Marion D, Franzetti B, Brutscher B, Boisbouvier J (2009) Fast two-dimensional NMR spectroscopy of high molecular weight protein assemblies. J Am Chem Soc 131:3448–3449
Amero C, Durá MA, Noirclerc-Savoye M, Perollier A, Gallet B, Plevin MJ, Vernet T, Franzetti B, Boisbouvier J (2011) A systematic mutagenesis-driven strategy for site-resolved NMR studies of supramolecular assemblies. J Biomol NMR 50:229–236
Ayala I, Sounier R, Usé N, Gans P, Boisbouvier J (2009) An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR 43:111–119
Ayala I, Hamelin O, Amero C, Pessey O, Plevin MJ, Gans P, Boisbouvier J (2012) An optimized isotopic labelling strategy of isoleucine-γ2 methyl groups for solution NMR studies of high molecular weight proteins. Chem Commun 48:1434–1436
Borissenko L, Groll M (2005) Crystal structure of TET protease reveals complementary protein degradation pathways in prokaryotes. J Mol Biol 346:1207–1219
De Carvalho LPS, Argyrou A, Blanchard JS (2005) Slow-onset feedback inhibition: α inhibition of mycobacterium tuberculosis α-isopropylmalate synthase by l-leucine. J Am Chem Soc 127:10004–10005
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Fischer M, Kloiber K, Häusler J, Ledolter K, Konrat R, Schmid W (2007) Synthesis of a 13C methyl group labeled methionine precursor as a useful tool for simplifying protein structural analysis by NMR spectroscopy. Chem Biochem 8:610–612
Gans P, Hamelin O, Sounier R, Ayala I, Durá MA, Amero CD, Noirclerc-Savoye M, Franzetti B, Plevin MJ, Boisbouvier J (2010) Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high molecular weight proteins. Ang Chem Int Ed 49:1958–1962
Gardner KH, Kay LE (1997) Production and incorporation of 15N, 13C, 2H (1H-δ1 methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc 119:7599–7600
Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG (2007) Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131:756–769
Godoy-Ruiz R, Guo C, Tugarinov V (2010) Alanine methyl groups as NMR probes of molecular structure and dynamics in high-molecular-weight proteins. J Am Chem Soc 132:18340–18350
Goto N, Gardner K, Mueller G, Willis R, Kay L (1999) A robust and cost-effective method for the production of Val, Leu, Ile (δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR 13:369–374
Gross JD, Gelev VM, Wagner G (2003) A sensitive and robust method for obtaining intermolecular NOEs between side chains in large protein complexes. J Biomol NMR 25:235–242
Hajduk PJ, Augeri DJ, Mack J, Mendoza R, Yang J, Betz SF, Fesik SW (2000) NMR-based screening of proteins containing 13C-labeled methyl groups. J Am Chem Soc 122:7898–7904
Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210
Howard BR, Endrizzi JA, Remington SJ (2000) Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 Å resolution: mechanistic implications. Biochemistry 39:3156–3168
Hu W, Namanja AT, Wong S, Chen Y (2012) Selective editing of Val and Leu methyl groups in high molecular weight protein NMR. J Biomol NMR 53:113–124
Isaacson RL, Simpson PJ, Liu M, Cota E, Zhang X, Freemont P, Matthews S (2007) A new labeling method for methyl transverse relaxation-optimized spectroscopy NMR spectra of alanine residues. J Am Chem Soc 129:15428–15429
Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei Ono A, Güntert P (2006) Optimal isotope labelling for NMR protein structure determinations. Nature 440:52–57
Lichtenecker R, Ludwiczek ML, Schmid W, Konrat R (2004) Simplification of protein NOESY spectra using bioorganic precursor synthesis and NMR spectral editing. J Am Chem Soc 126:5348–5349
Lichtenecker RJ, Coudevylle N, Konrat R, Schmid W (2013) Selective isotope labelling of leucine residues by using α-ketoacid precursor compounds. Chem Biochem 14:818–821
Miyanoiri Y, Takeda M, Okuma K, Ono AM, Terauchi T, Kainosho M (2013) Differential isotope-labeling for Leu and Val residues in a protein by E. coli cellular expression using stereo-specifically methyl labeled amino acids. J Biomol NMR. doi:10.1007/s10858-013-9784-0
Plevin MJ, Boisbouvier J (2012) Isotope-labelling of methyl groups for NMR studies of large proteins. In: Clore GM, Potts J (eds) Recent developments in biomolecular NMR. Royal Society of Chemistry 1–24. doi:10.1039/9781849735391-00001
Ruschak A, Kay L (2010) Methyl groups as probes of supra-molecular structure, dynamics and function. J Biomol NMR 46:75–87
Sounier R, Blanchard L, Wu Z, Boisbouvier J (2007) High-accuracy distance measurement between remote methyls in specifically protonated proteins. J Am Chem Soc 129:472–473
Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445:618–622
Sprangers R, Velyvis A, Kay LE (2007) Solution NMR of supramolecular complexes: providing new insights into function. Nat Meth 4:697–703
Tugarinov V, Kay LE (2003) Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc 125:13868–13878
Tugarinov V, Kay LE (2004) An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR 28:165–172
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced 1H–13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428
Tugarinov V, Choy W-Y, Orekhov VY, Kay LE (2005) Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc Natl Acad Sci USA 102:622–627
Tugarinov V, Kanelis V, Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc 1:749–754
Velyvis A, Ruschak AM, Kay LE (2012) An economical method for production of 2H, 13CH3-threonine for solution NMR studies of large protein complexes: application to the 670 kDa proteasome. PLoS One 7:e43725
Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696
Acknowledgments
We would like to thank Drs. M. Kainosho, P. Macek, M. Plevin, P. Schanda, A. Sivertsen, Mrs I. Ayala and R. Kerfah, as well as Mr. T. Ogden, for stimulating discussions, Dr. D. Marion for help in processing the NMR spectra and Drs. T. Vernet and M. Noirclerc-Savoye for the preparation of the library of mutants. We thank Dr. B. Franzetti for providing clones of TET2. This work used the RoBioMol, high-field NMR and the isotopic labeling facilities at the Grenoble Instruct Centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Program FP7/2007-2013 Grant Agreement no. 260887.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10858_2013_9785_MOESM1_ESM.pdf
The NMR spectra used to characterize the incorporation of exogenous l-leucine in overexpressed ubiquitin, as well as the table and the spectra corresponding to the assignment of the TET2 valine methyl groups, are available online: http://link.springer.com/journal/volumesAndIssues/10858 (PDF 2499 kb)
Rights and permissions
About this article
Cite this article
Mas, G., Crublet, E., Hamelin, O. et al. Specific labeling and assignment strategies of valine methyl groups for NMR studies of high molecular weight proteins. J Biomol NMR 57, 251–262 (2013). https://doi.org/10.1007/s10858-013-9785-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9785-z