Abstract
Purpose
Recurrent implantation failure (RIF) affects up to 10% of in vitro fertilization (IVF) patients worldwide. However, the pathogenesis of RIF remains unclear. This study was aimed at identifying hub transcription factors (TFs) of RIF in bioinformatics approaches.
Methods
The GSE111974 (mRNA), GSE71332 (miRNA), and GSE103465 (mRNA) datasets were downloaded from the Gene Expression Omnibus database from human endometrial tissue using R version 4.2.1 and used to identify differentially expressed TFs (DETFs), differentially expressed miRNAs, and differentially expressed genes for RIF, respectively. DETFs were subjected to functional enrichment analysis and the protein–protein interaction network analysis using the Search Tool for the Retrieval of Interacting Genes (version 11.5) database. Hub TFs were identified using the cytoHubb plug-in, after which a hub TF–miRNA–mRNA network was constructed using Cytoscape v3.8.2.
Results
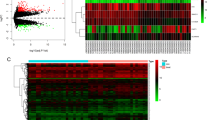
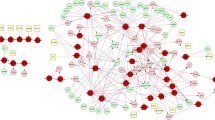
Fifty-seven DETFs were identified, in which Gene Ontology analysis revealed to be mainly involved in the regulation of transcription. Kyoto Encyclopedia of Genes and Genomes pathway analysis suggested that DETFs were enriched in transcriptional misregulation in cancer, aldosterone synthesis and secretion, AMPK signaling pathway, and cGMP-PKG signaling pathway. EOMES, NKX2-1, and POU5F1 were identified as hub TFs, and a hub TF–miRNA–mRNA regulatory network was constructed using these three hub TFs, four miRNAs, and four genes.
Conclusion
Collectively, we identified three promising molecular biomarkers for the diagnosis of RIF, which may further be potential therapeutic targets. This study provides novel insights into the molecular mechanisms underlying RIF. However, further experiments are required to verify these results.







Similar content being viewed by others
Data availability
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
References
Busnelli A, Reschini M, Cardellicchio L, Vegetti W, Somigliana E, Vercellini P. How common is real repeated implantation failure? An indirect estimate of the prevalence. Reprod Biomed Online. 2020;40(1):91–7. https://doi.org/10.1016/j.rbmo.2019.10.014.
Cimadomo D, Craciunas L, Vermeulen N, Vomstein K, Toth B. Definition, diagnostic and therapeutic options in recurrent implantation failure: an international survey of clinicians and embryologists. Hum Reprod. 2021;36(2):305–17. https://doi.org/10.1093/humrep/deaa317.
Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175(7):1842-55.e16. https://doi.org/10.1016/j.cell.2018.10.042.
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell. 2018;172(4):650–65. https://doi.org/10.1016/j.cell.2018.01.029.
Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1(1):31. https://doi.org/10.1186/2045-3701-1-31.
Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18(5):504–11. https://doi.org/10.1101/gad.1184404.
Shang J, Cheng YF, Li M, Wang H, Zhang JN, Guo XM, et al. Identification of key endometrial microRNAs and their target genes associated with pathogenesis of recurrent implantation failure by integrated bioinformatics analysis. Front Genet. 2022;13:919301. https://doi.org/10.3389/fgene.2022.919301.
Chen P, Li T, Guo Y, Jia L, Wang Y, Fang C. Construction of circulating microRNAs-based non-invasive prediction models of recurrent implantation failure by network analysis. Front Genet. 2021;12:712150. https://doi.org/10.3389/fgene.2021.712150.
Azhari F, Pence S, Hosseini MK, Balci BK, Cevik N, Bastu E, et al. The role of the serum exosomal and endometrial microRNAs in recurrent implantation failure. J Matern Fetal Neonatal Med. 2022;35(5):815–25. https://doi.org/10.1080/14767058.2020.1849095.
Luo J, Zhu L, Zhou N, Zhang Y, Zhang L, Zhang R. Construction of circular RNA-microRNA-messenger RNA regulatory network of recurrent implantation failure to explore its potential pathogenesis. Front Genet. 2020;11:627459. https://doi.org/10.3389/fgene.2020.627459.
Ahmadi M, Pashangzadeh S, Moraghebi M, Sabetian S, Shekari M, Eini F, et al. Construction of circRNA-miRNA-mRNA network in the pathogenesis of recurrent implantation failure using integrated bioinformatics study. J Cell Mol Med. 2022;26(6):1853–64. https://doi.org/10.1111/jcmm.16586.
Bastu E, Demiral I, Gunel T, Ulgen E, Gumusoglu E, Hosseini MK, et al. Potential marker pathways in the endometrium that may cause recurrent implantation failure. Reprod Sci. 2019;26(7):879–90. https://doi.org/10.1177/1933719118792104.
Shi C, Shen H, Fan LJ, Guan J, Zheng XB, Chen X, et al. Endometrial microRNA signature during the window of implantation changed in patients with repeated implantation failure. Chin Med J (Engl). 2017;130(5):566–73. https://doi.org/10.4103/0366-6999.200550.
Guo F, Si C, Zhou M, Wang J, Zhang D, Leung PCK, et al. Decreased PECAM1-mediated TGF-beta1 expression in the mid-secretory endometrium in women with recurrent implantation failure. Hum Reprod. 2018;33(5):832–43. https://doi.org/10.1093/humrep/dey022.
Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47(D1):D33–8. https://doi.org/10.1093/nar/gky822.
Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158(6):1431–43. https://doi.org/10.1016/j.cell.2014.08.009.
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–46. https://doi.org/10.1093/nar/gkac1000.
Tong Z, Cui Q, Wang J, Zhou Y. TransmiR v2.0: an updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2019;47(D1):D253–8. https://doi.org/10.1093/nar/gky1023.
Montaldo E, Vacca P, Chiossone L, Croxatto D, Loiacono F, Martini S, et al. Unique Eomes(+) NK cell subsets are present in uterus and decidua during early pregnancy. Front Immunol. 2015;6:646. https://doi.org/10.3389/fimmu.2015.00646.
Kiekens L, Van Loocke W, Taveirne S, Wahlen S, Persyn E, Van Ammel E, et al. T-BET and EOMES accelerate and enhance functional differentiation of human natural killer cells. Front Immunol. 2021;12:732511. https://doi.org/10.3389/fimmu.2021.732511.
Chen L, Li M, Sun F, Qian J, Du M, Wang S, et al. Eomesodermin regulate decidual CD4(+)T cell function during human early pregnancy. J Reprod Immunol. 2021;146:103290. https://doi.org/10.1016/j.jri.2021.103290.
Chen L, Sun F, Li M, Qian J, Du M, Li D, et al. Decreased level of Eomes+dCD8+ T cells with altered function might be associated with miscarriage. Reproduction. 2021;162(2):107–15. https://doi.org/10.1530/REP-20-0639.
Probst S, Arnold SJ. Eomesodermin-at dawn of cell fate decisions during early embryogenesis. Curr Top Dev Biol. 2017;122:93–115. https://doi.org/10.1016/bs.ctdb.2016.09.001.
Lai ZZ, Wang Y, Zhou WJ, Liang Z, Shi JW, Yang HL, et al. Single-cell transcriptome profiling of the human endometrium of patients with recurrent implantation failure. Theranostics. 2022;12(15):6527–47. https://doi.org/10.7150/thno.74053.
Bakhmet EI, Tomilin AN. Key features of the POU transcription factor Oct4 from an evolutionary perspective. Cell Mol Life Sci. 2021;78(23):7339–53. https://doi.org/10.1007/s00018-021-03975-8.
Hu J, Yuan R. Decreased expression of c-kit and telomerase in a rat model of chronic endometrial ischemia. Med Sci Monit. 2011;17(4):BR103-9. https://doi.org/10.12659/msm.881710.
Liu H, Deng S, Zhao Z, Zhang H, Xiao J, Song W, et al. Oct4 regulates the miR-302 cluster in P19 mouse embryonic carcinoma cells. Mol Biol Rep. 2011;38(3):2155–60. https://doi.org/10.1007/s11033-010-0343-4.
Wang YD, Cai N, Wu XL, Cao HZ, Xie LL, Zheng PS. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013;4:e760. https://doi.org/10.1038/cddis.2013.272.
Minocha S, Valloton D, Arsenijevic Y, Cardinaux JR, Guidi R, Hornung JP, et al. Nkx2.1 regulates the generation of telencephalic astrocytes during embryonic development. Sci Rep. 2017;7:43093. https://doi.org/10.1038/srep43093.
Wen B, Li E, Ustiyan V, Wang G, Guo M, Na CL, et al. In vivo generation of lung and thyroid tissues from embryonic stem cells using blastocyst complementation. Am J Respir Crit Care Med. 2021;203(4):471–83. https://doi.org/10.1164/rccm.201909-1836OC.
Tagne JB, Mohtar OR, Campbell JD, Lakshminarayanan M, Huang J, Hinds AC, et al. Transcription factor and microRNA interactions in lung cells: an inhibitory link between NK2 homeobox 1, miR-200c and the developmental and oncogenic factors Nfib and Myb. Respir Res. 2015;16:22. https://doi.org/10.1186/s12931-015-0186-6.
Ponsuksili S, Tesfaye D, Schellander K, Hoelker M, Hadlich F, Schwerin M, et al. Differential expression of miRNAs and their target mRNAs in endometria prior to maternal recognition of pregnancy associates with endometrial receptivity for in vivo- and in vitro-produced bovine embryos. Biol Reprod. 2014;91(6):135. https://doi.org/10.1095/biolreprod.114.121392.
Shi S, Tan Q, Liang J, Cao D, Wang S, Liang J, et al. Placental trophoblast cell-derived exosomal microRNA-1290 promotes the interaction between endometrium and embryo by targeting LHX6. Mol Ther Nucleic Acids. 2021;26:760–72. https://doi.org/10.1016/j.omtn.2021.09.009.
Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, et al. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol. 2009;334(1):186–97. https://doi.org/10.1016/j.ydbio.2009.07.016.
Al-Ashtal HA, Rubottom CM, Leeper TC, Berman AJ. The LARP1 La-Module recognizes both ends of TOP mRNAs. RNA Biol. 2021;18(2):248–58. https://doi.org/10.1080/15476286.2019.1669404.
Hug C, Miller TM, Torres MA, Casella JF, Cooper JA. Identification and characterization of an actin-binding site of CapZ. J Cell Biol. 1992;116(4):923–31. https://doi.org/10.1083/jcb.116.4.923.
Alshaker H, Wang Q, Brewer D, Pchejetski D. Transcriptome-wide effects of sphingosine kinases knockdown in metastatic prostate and breast cancer cells: implications for therapeutic targeting. Front Pharmacol. 2019;10:303. https://doi.org/10.3389/fphar.2019.00303.
Kim SJ, Lee SY, Lee C, Kim I, An HJ, Kim JY, et al. Differential expression profiling of genes in a complete hydatidiform mole using cDNA microarray analysis. Gynecol Oncol. 2006;103(2):654–60. https://doi.org/10.1016/j.ygyno.2006.05.015.
Martinsson-Ahlzen HS, Liberal V, Grunenfelder B, Chaves SR, Spruck CH, Reed SI. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol. 2008;28(18):5698–709. https://doi.org/10.1128/MCB.01833-07.
Ellederova Z, Del Rincon S, Koncicka M, Susor A, Kubelka M, Sun D, et al. CKS1 germ line exclusion is essential for the transition from meiosis to early embryonic development. Mol Cell Biol. 2019;39(13). https://doi.org/10.1128/MCB.00590-18
Kang MA, Kim JT, Kim JH, Kim SY, Kim YH, Yeom YI, et al. Upregulation of the cycline kinase subunit CKS2 increases cell proliferation rate in gastric cancer. J Cancer Res Clin Oncol. 2009;135(6):761–9. https://doi.org/10.1007/s00432-008-0510-3.
Shen DY, Zhan YH, Wang QM, Rui G, Zhang ZM. Oncogenic potential of cyclin kinase subunit-2 in cholangiocarcinoma. Liver Int. 2013;33(1):137–48. https://doi.org/10.1111/liv.12014.
Huberts DH, van der Klei IJ. Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta. 2010;1803(4):520–5. https://doi.org/10.1016/j.bbamcr.2010.01.022.
Liu J, Wang Y, Chen P, Ma Y, Wang S, Tian Y, et al. AC002454 1 and CDK6 synergistically promote endometrial cell migration and invasion in endometriosis. Reproduction. 2019;157(6):535–43. https://doi.org/10.1530/REP-19-0005.
Funding
This manuscript was supported by the National Natural Science Foundation of China (grant number 81860271 and 82160281), Yunnan Ten Thousand Talents Program for Famous Doctors and Masters (grant number Yunwei Renfa [2019] No. 35), Yunnan Support Program of High Level Talents Cultivation Famous Medical Project (grant number RLMY20200017), Fundamental Research Joint Project of Local Undergraduate Universities in Yunnan Province (grant number 2020D1BA07000), Yunnan Provincial Clinical Medical Center for Reproductive Obstetrics and Gynecology (grant number zx209-01-01), Open Subject of Yunnan Clinical Medical Center for Reproductive and Maternal Diseases (grant number 2019LCZXKF-SZ04), Dali Science and Technology Planning Project (grant numbers 2021085 and 2021086), and Open Program of Clinical Medical Center of First People’s Hospital of Yunnan Province (2022LCZXKF-SZ13).
Author information
Authors and Affiliations
Contributions
Conceptualization: JL, RH, and PX. Formal analysis: AX, ZD, LZ, RW, YQ, and LZ. Methodology: RH, PX, AX, ZD, LZ, RW, YQ, and LZ. Software: JL, AX, and ZD. Supervision: RZ and LT. Writing—original draft, JL, RH, and PX. Writing—review and editing: LZ, RZ, and LT. Project administration: RZ and LT. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Publicly available datasets were analyzed in this study, so ethical review, approval, and written informed consent for participation were not required for the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, J., Huang, R., Xiao, P. et al. Construction of hub transcription factor–microRNAs–messenger RNA regulatory network in recurrent implantation failure. J Assist Reprod Genet 41, 3–13 (2024). https://doi.org/10.1007/s10815-023-02947-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02947-0