Abstract
Microalgae cultivation on wastewater offers the dual benefit of lowering costs for feedstock production with simultaneous wastewater remediation. This study utilized biochemical and quantitative label-free proteomic approaches to evaluate the growth and proteomic response for diatom Phaeodactylum tricornutum cultivated on flushed dairy manure wastewater (DMW). Comparing several DMW dilutions (up to 60% DMW diluted in seawater) with a synthetic seawater medium indicates that biomass and lipid yields correlate with the starting nitrogen content of the DMW dilution. Phaeodactylum tricornutum cultivated on DMW exhibits elevated levels of polyunsaturated fatty acids (PUFAs), particularly docosapentaenoic acid (DPA, 22:5 n-3). Proteomic analysis revealed alterations in the regulations of proteins associated with protein metabolism, cellular signaling, transcription and translation, protein trafficking, and oxidative stress management pathways when comparing P. tricornutum cultivation on diluted DMW versus synthetic media, thus providing insights into how P. tricornutum reorganizes its proteome in response to a complex wastewater source.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Microalgal lipids are a promising renewable feedstock for the production of biodiesel and other high value co-products. Microalgae can grow in aquatic environments with varying salinities, possess higher growth and overall lipid production rates when compared to their terrestrial biofuel feedstock crop counterparts, and avoid conflicts over the use of terrestrial resources for fuel versus food production (Hannon et al. 2010). Despite these advantages, algae-derived biofuels are currently not competitive compared to fossil fuels, partly due to the high costs associated with nutrient and water use for cultivating algae biomass (Fargione et al. 2008). Generating 1 kg of algae-derived biofuels requires 3000 kg of water, 0.33 kg of nitrogen, and 0.71 kg of phosphate when nutrients and water are not recycled (Jiang et al. 2011). The large nutrient and water input required for microalgae-based biofuel production suggest the importance of finding alternative nutrient and water sources for microalgae cultivation.
The use of wastewater sources such as flushed dairy manure wastewater (DMW) for algal biomass production is a viable strategy for reducing microalgae cultivation costs while also having potential benefit to the dairy industry (Chokshi et al. 2016). Microalgae can utilize inorganic nutrients present in DMW for growth and cellular processes, with nitrogen and phosphorus concentrations of DMW reaching 9.2–24.3 g kg-1 and 1.2–4.1 g kg-1 dry weight, respectively (Moreno-Caselles et al. 2002). With over 20 million tonnes per year of dairy manure produced yearly by dairy farms nationwide (Smith et al. 2016), unused manure must be managed to reduce negative environmental impacts such as surface water eutrophication and aquifer contamination. In general, DMW is applied as a crop fertilizer and in lesser amounts as a substrate for biogas production via anaerobic digestion (Demirer and Chen 2005). Additional opportunities exist to further exploit the use of DMW as an alternate nutrient source for microalgae biomass production.
Previous reports have demonstrated successful cultivation of microalgae species on a variety of wastewater sources (for review, see Rawat et al. 2016), but little is known about the molecular responses associated with microalgae cultivation in wastewater. “Omics” technologies represent an important option to investigate how microalgae respond to various wastewater cultivation practices, yet there is only one report of an “omic” approach to investigate microalgae responses to cultivation on wastewater. Patel and coworkers have reported the use of MS-based proteomics to characterize changes in the proteome of Chlamydomonas reinhardtii following cultivation on a synthetic wastewater medium (Patel et al. 2015). Previously, the use of “omic” technologies has also been employed to investigate microalgae responses to various alterations in environmental conditions, including nitrogen deprivation (Park et al. 2015), presence of heavy metals (Simon et al. 2013), and high saline concentrations (Ho et al. 2017).
Herein, we present the first example using proteomic analysis for evaluating the molecular response of microalgae cultured on an authentic (non-artificial) wastewater source. We use label-free quantitative proteomics combined with biochemical and physiological analyses to investigate the response of the marine diatom Phaeodactylum tricornutum cultivated on a seawater-diluted flushed DMW source, first by determining optimal DMW concentrations for comparison to synthetic media and then employing proteomics. Phaeodactylum tricornutum was selected for this study based on its high lipid content, sequenced and annotated reference genome (Bowler et al. 2008; Michele Fabris et al. 2012), and documented ability to grow on wastewater sources (Coppens et al. 2014; Libralato et al. 2016). This study highlights the utility of DMW as an alternative nutrient source for microalgae cultivation and provides insights into how P. tricornutum reorganizes its proteome in response to cultivation on a complex wastewater source.
Material and methods
Flushed dairy manure wastewater collection, filtration, and characterization
Flushed DMW was collected from a manure storage lagoon located in Merced County of California, USA. Prior to entering the storage lagoon, flushed dairy manure was passed through a solid separator, which separates flushed manure into liquid and solid manure streams. The liquid manure stream was stored in a lagoon. Once collected from the lagoon and transported, the DMW was stored at 4 °C prior to use. Prior to its preparation as a culturing medium, the DMW was passed through a Wypall filter (Kimberly Clark, USA) and centrifuged to remove large suspended solids, producing a coarse-filtered DMW solution. Coarse-filtered DMW was subsequently sterilized by filtering through a 0.22-μm filter (Merck Millipore, USA) to produce the sterile-filtered DMW solution that was used throughout the study. Samples of both unfiltered and sterile-filtered DMW were analyzed by Denele Analytical, Inc. (Woodland, California, USA) using their Complete Lagoon Water Analysis package. For use as a culturing medium, DMW was diluted to specified dilutions using autoclaved, sterile-filtered seawater collected from the UC Davis-Bodega Marine Laboratory (Bodega Bay, CA, USA).
Microalgae culture maintenance on synthetic medium
An axenic culture of P. tricornutum (UTEX 466) was obtained from UTEX (University of Texas, Austin, TX, USA) and maintained on agar plates made of f/2 (F2) medium (NCMA, East Boothbay, ME, USA). Prior to inoculation into DMW dilutions as growth medium, P. tricornutum colonies from agar plates were inoculated into F2 liquid culture in 500-mL Corning Pyrex media bottles (Corning 1395-2 L) using stir bars with air bubbling (Petco Air Pump). During algae cultivation, incoming air was sterile-filtered by Polyvent 4 disposable filters (Whatman, UK). Culture suspensions were maintained at a constant temperature of 23 ± 2 °C with full spectrum incident uniform lighting (High Efficiency T-5 Grow Lights, Gardeners Supply Co, VT, USA) at a 16:8 h light/dark cycle (60–120 μmol photons m-2 s-1). All equipment and materials were autoclaved and media added to flasks in a Labconco Purifier Vertical Clean Bench (Labconco, USA) using sterile technique practices. Cell density was measured every other day at 680 nm using a Thermo Scientific Genesys 10S Vis Spectrophotometer (Thermo Fisher, USA).
Microalgae cultivation on sterile-filtered dairy manure wastewater
Liquid stock cultures of P. tricornutum were grown to a mid-logarithmic growth phase based on absorbance measurements at 680 nm. Prior to inoculation, cell counts on algae stock cultures were performed using an Improved Neubauer Hemocytometer (Hausser Scientific, USA) and inverted microscope (Eclipse TS100, Nikon, Japan). Algae stock solutions were inoculated into DMW dilutions at a 1:9 inoculant/DMW dilution ratio. All experiments were performed using 3–5 biological replicates and compared to control cultures inoculated into synthetic medium (F2). Throughout the experiment, growth was monitored by cell counting as described above.
Chlorophyll a determination
The chlorophyll a content of samples was measured by extraction using similar methods described by Ritchie (2006). Briefly, 1.0 mL of culture volumes was pelleted by centrifugation, washed with distilled H2O (dH2O), and re-pelleted. Following removal of the supernatant, microalgae pellets were flash frozen in liquid nitrogen and extracted in 1 mL of ice-cold methanol (100%, ACS grade). Chlorophyll a content was determined spectrophotometrically using the equation: chlorophyll a (μg mL-1) = 12.9447x(Abs665-Abs720).
Biomass determination
At the end of the experiment, microalgae cultures (250–300 mL) were pelleted by centrifugation and washed once with dH2O. Algae pellets were then frozen in liquid nitrogen and lyophilized. Lyophilized biomass weights were recorded and are reported as milligram of harvested biomass per liter culture (mg L-1). Lyophilized biomass was stored at – 20 °C under argon gas until utilized for lipid extraction assays as described below.
Lipid extraction
Total lipids were extracted from freeze-dried algae biomass using a modified Folch method (Folch et al. 1957). Microalgae samples were first treated with chloroform and pulverized using a hand sonicator (Fisher Scientific Model 120 Sonic Dismembrator, Thermo Fisher, USA) for 1 min. Next, a 2:1 mixture of methanol/chloroform was added to samples and sonication repeated. Then, 0.1 M PBS buffer was added to samples, mixed vigorously, and centrifuged to separate layers. The lower organic (chloroform) phase containing lipids was removed and stored at – 20 °C.
Neutral lipid content determination using a Nile Red live cell assay
Intracellular lipid analysis of live P. tricornutum cultures was performed in a 96-well microplate as described by Franz and colleagues with slight modifications (Franz et al. 2013). Briefly, 25 μL of a 1:1 (v/v) DMSO/filtered seawater solution was added to 250 μL P. tricornutum, followed by addition of 4 μL of 1 mg mL-1 Nile Red (9-diethylamino-5H-benzo[α]phenoxazine-5-one, Sigma, USA) dissolved in acetone. Following a 5-min incubation in the dark, fluorescence measurements were captured using a Synergy HI Multi-Mode Plate Reader (Biotek, USA) at excitation and emission wavelengths of 530/40 nm and 590/40 nm, respectively. Background fluorescent measurements obtained from DMW dilution blanks were subtracted from wells prior to analysis.
Neutral lipid content determination using a bleach-enhanced Nile Red microplate lipid extract assay
A bleach-enhanced Nile Red microplate assay was adapted and used to quantify the amount of neutral lipids in the total lipid extract from microalgae (Higgins et al. 2014). This quantitative assay is referred to as a Nile Red neutral lipid assay and has been previously described by Burch and Franz (2016).
Lipid extraction and conversion to fatty acid methyl esters (FAMEs)
Lyophilized P. tricornutum biomass (40–50 mg) samples were spiked with 1 μmol of an internal standard, heptadecanoic acid (Nu-check Prep, Inc., USA) dissolved in 100 μL of chloroform. To each sample, 400 μL of toluene, 3 mL of methanol, and 600 μL of 8% HCl in methanol solution were added. Samples were capped and incubated at 90 °C for 60 min and then allowed to cool for 10 min. After cooling, 1 mL of hexane and then 1 mL of water were added to each sample to quench the reaction. Samples were vortexed and allowed to separate into organic/aqueous layers for 15 min, and complete separation of layers was attained by centrifugation. The upper hexane layer was withdrawn and washed with water, and then the remaining cellular debris was removed by centrifugation. FAME samples were stored at – 80 °C until analysis (Ichihara and Fukubayashi 2010).
Analysis of FAMEs by gas chromatography
FAMEs were analyzed using an Agilent 3800 gas chromatography system with a DB-23 fused silica column (0.25 mm ID × 30 m L, 0.25 mm film thickness; Agilent). Oven temperature was initially held at 50 °C for 2 min, ramped at 10 °C min-1 to 180 °C, held for 5 min, ramped to 240 °C at 5 °C min-1, and held for 5 min. Injector and detector temperatures were held at 250 °C and 300 °C, respectively. Helium was used as the carrier gas (1.3 mL min-1) and inlet makeup gas. FAMEs were identified by retention time using a mix of 31 FAME standards (NuChek Prep, Inc., USA), and FAME quantification was calculated by peak area relative to the internal standard as previously described (Ichihara and Fukubayashi 2010). Unidentified peaks were excluded from analysis.
Proteomic sample preparation
Phaeodactylum tricornutum samples were collected for label-free quantitative proteomic following 5 days of cultivation on DMW dilutions or F2 medium. Briefly, 10 mL of algae was pelleted by centrifugation and washed with sterile-filtered seawater. Following a second centrifugation step, washed algae pellets were flash frozen in liquid nitrogen and stored at – 80 °C until processed for proteomic analysis. Five biological replicates per condition (synthetic medium, 10% DMW, 20% DMW) were used for proteomic analysis (n = 5).
Thawed algae pellets were resuspended in RIPA buffer (Thermo Fisher Scientific, USA) containing protease inhibitors (Roche cOmplete ULTRA Tablets, Sigma Aldrich, USA) and lysed by mechanical disruption using a MagNA Lyser (Roche Diagnostics, USA). A 100 μg of protein from each sample was precipitated using Calbiochem ProteoExtract Protein Precipitation Kit (Merck Millipore, USA). Precipitated proteins were resuspended into a 50 mM NH3HCO3 buffer containing 6.0 M urea and then reduced and subsequently alkylated using dithiothreitol (DTT) and iodoacetamide (IAA), respectively. Enzymatic digestion of proteins was performed using MS-grade LysC (Wako Chemicals USA) for 4 h at 37 °C and then diluted to reduce urea concentration and allowed to further digest overnight with sequencing grade modified trypsin (Promega, USA). After digestion, protein samples were desalted using a Macrospin microSPE kit (Nest Group, USA).
Digested peptide samples were analyzed by LC-MS/MS at the UC Davis Proteomic Core Facility using a Thermo Scientific Q Exactive Plus Orbitrap Mass Spectrometer in conjunction with a Proxeon Easy-nLC II HPLC and Proxeon nanospray source. The digested peptides were loaded on a 100 μm × 25 mm Magic C18 100 Å 5 U reverse phase trap where they were desalted online before being separated using a 75 μm × 150 mm Magic C18 200 Å 3 U reverse phase column. Peptides were eluted using a 120-min gradient (2–35% buffer B for 85 min, 35–80% buffer B for 25 min, 2% buffer B for 10 min; buffer A = 0.01% formic acid in H2O, buffer B = 100% acetonitrile) with a flow rate of 300 nL min-1. An MS survey scan was obtained for the m/z range 300–1600, and MS/MS spectra were acquired using a top 15 method, where the top 15 ions in the MS spectra were subjected to high energy collisional dissociation (HCD). An isolation mass window of 1.6 m/z was used for the precursor ion selection, with a normalized collision energy of 27% for fragmentation. Fifteen seconds was used for the dynamic exclusion.
Proteomic data analysis
Peptide spectral matching (PSM) and protein identification was performed using MaxQuant (v. 1.5.7.4) against the UniProt reference proteome for P. tricornutum with the default MaxQuant laboratory contaminant sequences (Apweiler et al. 2004). Decoy false discovery rates (FDR) were limited to less than 1% at the peptide and protein level. Identified proteins were considered for subsequent downstream analysis only if they were identified in at least three biological replicates in at least one experimental condition. The Perseus (v. 1.5.6.0) computation platform was used for statistical analysis, and a one-way ANOVA was performed using a permutation-based FDR strategy (FDR > 0.05) at a significance level of 5%. Subsequent post hoc analysis (Tukey’s HSD) and two-tailed t-tests were performed to identify and determine fold change differences in significant (p < 0.05) proteins across conditions (F2, 10% DMW, 20% DMW). Proteins were utilized in downstream analysis only if the observed fold change between conditions was greater than 2-fold (log2FC ± 1) and p value < 0.05. In this study, in-text fold changes (log2FC ± 1 and p value < 0.05) are reported after UniProt IDs (e.g., UniProt ID, 10%/20% + 2.0), with the percentage values indicating the conditions being compared. Functional annotations, including gene ontology (GO), pfam, and interpro annotations for each observed significant protein, were obtained from the UniProt website. Additional annotation was performed using the Mercator webserver (Lohse et al. 2014) followed by manual curation using DiatomCyc (Fabris et al. 2012), KEGG (Kanehisa and Goto 2000), and JGI databases (Nordberg et al. 2013). Visualization of data was performed using MapMan software (Thimm et al. 2004), the KEGG Mapper “Search&Color Pathway” tool (Kanehisa et al. 2012), and JColorGrid (Joachimiak et al. 2006).
Microscopy evaluation of P. tricornutum grown on dairy manure wastewater
Microscopic analysis and imaging of P. tricornutum grown on F2 and DMW dilutions were performed at multiple time points throughout the culture period using an inverted light microscope (Nikon TS100 Eclipse). Images were acquired using a mounted camera (Canon EOS Rebel XS) in combination with a × 100 oil immersion lens (Nikon CFI Achro 100X 1.4 NA). Images were processed using ImageJ version 1.52a (Schneider et al. 2012).
Total nitrogen content analysis
The total nitrogen content of F2 and DMW media sources were determined by a persulfate digestion method using a Hach Total Nitrogen Test N’ Tube Reagent Set (Hach, USA) according to the manufacturer’s recommendations.
Iron analysis of F2 and DMW media sources
Iron content of F2 and DMW media sources were determined by the UC Davis Analytical Laboratory using a nitric acid/hydrogen peroxide microwave digestion procedure followed by determination via atomic absorption spectrometry and inductively coupled plasma (ICP) atomic emission spectrometry.
Results
Analysis of dairy wastewater (DMW) composition
Synthetic media (F2), DMW, and sterile-filtered DMW were analyzed by Denele Analytical, Inc. (Woodland, California) using the Complete Lagoon Water Analysis package (Table 1). Sterile filtration of the DMW resulted in a notable reduction of total Kjeldahl nitrogen (TKN) from 123 to 56 mg L-1 and phosphorous from 21.7 to 4.57 mg L-1, resulting from volatilization and suspended solid exclusion respectively.
Cell density, biomass production, and chlorophyll a content of P. tricornutum cultivated using DMW
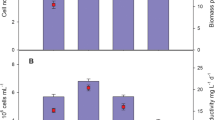
Phaeodactylum tricornutum cultivated on 10%, 30%, and 60% DMW for 15 days resulted in differences in cell density, biomass production, and chlorophyll a content when comparing DMW dilutions to each other and when compared to cultivation using synthetic medium (F2) over the same time period (Fig. 1a–c). Cell density after 15 days of culturing was similar in the F2, 30%, and 60% conditions, with all three conditions reaching between 4.97 × 106 and 5.24 × 106 cells mL-1. Compared to the F2 control, increases in the chlorophyll a content were observed using 30% and 60% DMW conditions (Fig. 1b). In both the F2 and 30% DMW conditions, chlorophyll a content peaked on day 10 at 2.6 μg mL-1 and 3.3 μg mL-1, respectively, and declined to 2.0 μg mL-1 and 2.7 μg mL-1 by day 15, respectively.
a Growth of P. tricornutum on synthetic medium (F2) or 10%, 30%, and 60% DMW dilutions over a 15-day period as measured by cell density, b chlorophyll a content, and c total harvested biomass (day 15). c Comparison of total biomass harvested on day 15 (primary y-axis) compared to the starting nitrogen content of diluted DMW versus synthetic medium (secondary y-axis). Data shown (a–c) as mean ± SD (n = 3). d Microscopic images (× 100) of P. tricornutum cultured on F2 or DMW dilutions on day 15 of the 15-day growth period
No decrease in chlorophyll a content was observed through the growth period in the 60% DMW condition, with the chlorophyll a content reaching 4.6 μg mL-1 by day 15. A similar pattern was observed in biomass harvested at day 15 (Fig. 1c), with F2, 30%, and 60% DMW yielding approximately 309 mg L-1, 569 mg L-1, and 774 mg L-1 dried biomass, respectively. A reduction in cell density was observed in the 10% DMW condition by day 15 compared to all other conditions, with cultures reaching approximately 2.34 × 106 cells mL-1. Similarly, P. tricornutum cultivated using 10% DMW had a lower chlorophyll a content throughout the culture period, peaking at 1.0 μg chlorophyll a per mL at day 3, with less biomass (247 mg L-1) harvested by day 15 when compared to all other conditions.
Neutral lipid content of P. tricornutum grown on DMW dilutions
Differences in the neutral lipid content of P. tricornutum grown on 10%, 30%, and 60% were observed when compared to cultures grown on F2 medium using two Nile Red staining methods (Fig. 2a,b). The differences in neutral lipid content of harvested algae biomass were confirmed at day 15 using an additional Nile Red assay (Burch and Franz 2016; Higgins et al. 2014) that compared the Nile Red fluorescence of lipid extracts against a known oil standard (Fig. 2b). An increase in the neutral lipid content was observed in the 10% DMW condition (0.36 mg neutral lipids per mg biomass (mg mg-1)) when compared to synthetic medium (F2) control (0.15 mg mg-1), 30% DMW (0.10 mg mg-1), and 60% DMW (0.034 mg mg-1).
a Neutral lipid content of P. tricornutum grown on synthetic medium (F2) or 10%, 30%, and 60% DMW dilutions as measured by in vivo Nile Red fluorescence. b Comparison of the neutral lipid content per biomass (mg mg-1) of day 15 crude lipid extracts (primary y-axis) compared to the starting nitrogen content of diluted DMW versus synthetic medium (secondary y-axis). Data shown as mean ± SD (n = 3)
Fatty acid profile of P. tricornutum grown on DMW dilutions
Cultivation of P. tricornutum on increasing concentrations of DMW generally decreased the levels of saturated fatty acids (SFAs) and monounsaturated fatty acids (MUFAs) while increasing docosapentaenoic acid (DPA, C22:5 n-6) relative to cultivation in synthetic medium (Table 2, Supplementary Table 1). Cultivation using 30% and 60% DMW shifted the fatty acid profile where an increase in DPA content was especially notable compared to F2 and 10% DMW conditions.
Overview of proteomic data comparing P. tricornutum growth on DMW dilutions to growth on synthetic medium
Phaeodactylum tricornutum was grown on either 10% DMW (nitrogen-limited), 20% DMW (nitrogen-equivalent), or synthetic medium (F2) and sampled for proteomic analysis after 5 days of cultivation. Using a label-free quantitative approach, a total of 1562 proteins were identified across all conditions, and an analysis of variance (ANOVA) test identified 268 proteins with significantly different (p < 0.05) expression patterns across all conditions. Post hoc analysis (Tukey’s HSD) and pairwise comparison of significantly different proteins across conditions (10% DMW-F2, 20% DMW-F2, and 10% DMW-20% DMW) identified 198, 104, and 102 proteins with significantly different expression across conditions, respectively (Fig. 3a,b). Fold change expression of significantly (p < 0.05, log2FC ± 1) regulated proteins compared to each condition are reflected in Fig. 4 and categorized into nitrogen metabolism (Fig. 4a), iron limitation (Fig. 4b), photosynthesis-related pathways (Fig. 4c), central carbon metabolism (Fig. 4d), redox-related pathways (Fig. 4e), and gene expression and signaling (Fig. 4f).
a Volcano plots of proteins showing fold change and statistical significance of change across comparison conditions. Significantly (p < 0.05, log2FC ± 1) regulated proteins are represented by red triangles in volcano plots. b Bar charts of proteins showing significant upregulation (green) and downregulation (red) across comparison conditions. **Post hoc comparisons using Tukey’s HSD test followed by independent t-tests where log2FC ± 1
Fold change expression of significantly (p < 0.05, log2FC ± 1) regulated proteins involved in a nitrogen metabolism, b iron limitation, c photosynthesis-related pathways, d central carbon metabolism, e redox-related pathways, and f gene expression and signaling. Rows represent proteins with unique UniProt ID identifier, and columns specify comparison condition. Blue and red colored boxes signify upregulation or downregulation of abundance in each specific comparison, respectively. Grey boxes represent no significant difference across comparison conditions
Discussion
Cell density, biomass production and chlorophyll a content of P. tricornutum cultivated using DMW
The difference in P. tricornutum harvested biomass and chlorophyll a content observed across conditions is attributed to variation in the nitrogen content of the diluted DMW and synthetic media sources used throughout the study. The 10%, 30%, and 60% DMW dilutions contain approximately -49%, 35%, and 244% of the nitrogen present in the synthetic medium, respectively (Fig. 1c). A positive relationship between nitrogen content, chlorophyll content, and growth has been observed across all major evolutionary lineages of microalgae including P. tricornutum and other diatom species (Qiao et al. 2016; Yodsuwan et al. 2017). Nitrogen limitation reduces the cellular chlorophyll content in P. tricornutum (Valenzuela et al. 2013) and is considered part of a nitrogen-recycling mechanism that repurposes the nitrogen contained within the chlorophyll porphyrin rings and chlorophyll binding proteins for other cellular processes (Hörtensteiner and Kräutler 2011). Discrepancies observed between cell density, chlorophyll a content, and biomass measurements (Fig. 1a–c) in the 30% and 60% DMW conditions compared to the F2 control may be in part due to the complex composition of the dairy wastewater medium source. The increase in biomass and chlorophyll a content observed following cultivation on 30% and 60% DMW dilutions is consistent with the higher starting nitrogen content in these conditions compared to the F2 control, yet no difference in the final cell density was observed at the end of the culture period. Microscopic observation of cultures throughout the culture period (Fig. 1d) rules out microbial contamination but revealed the presence of cellular debris in the 30% and 60% DMW conditions compared to the F2 and 10% DMW conditions. Therefore, these results present the possibility that an unidentified component of the wastewater may promote cell degradation in the higher DMW concentrations. These results informed our design of the proteomic experiments to include a “nitrogen equivalent” dilution of DMW compared to the F2 media (20%), as well as a “nitrogen deficient” dilution (10%).
Neutral lipid content of P. tricornutum grown on DMW dilutions
Differences in the neutral lipid content of P. tricornutum grown on 10%, 30%, and 60% were observed when compared to cultures grown on F2 medium using two Nile Red staining methods (Fig. 2a,b). The first Nile Red method (Chen et al. 2009; Franz et al. 2013) measured the relative intracellular lipid content throughout the 15-day culture period and revealed an increase in the relative intracellular lipid content of 10% DMW compared to all other conditions by day 10 (Fig. 2a). The neutral lipid content of P. tricornutum cultured in the 60% DMW condition remained relatively unchanged over time, whereas the neutral lipid content gradually increased throughout the culture period in the F2, 10%, and 30% DMW conditions. An increase in the neutral lipid content was observed in the 10% DMW condition (0.36 mg neutral lipids/mg biomass (mg mg-1)) when compared to synthetic medium (F2) control (0.15 mg mg-1), 30% DMW (0.10 mg mg-1), and 60% DMW (0.034 mg mg-1) (Fig. 2b). The increase in neutral lipid content following cultivation on the 10% DMW condition is most likely a response to nitrogen depletion conditions (Burch and Franz 2016; Yang et al. 2014; Yodsuwan et al. 2017) and is supported by the differences in growth, biomass, chlorophyll a content, and initial nitrogen concentrations detected between the conditions tested (Fig. 1a–c, Fig. 2b).
Fatty acid profile of P. tricornutum grown on DMW dilutions
Cultivation of P. tricornutum using more DMW generally decreased the levels of saturated fatty acids (SFAs) and monounsaturated fatty acids (MUFAs) while increasing docosapentaenoic acid (DPA, C22:5 n-6) relative to cultivation in synthetic medium (Table 2, Supplementary Table 1). Consistent with previous reports for P. tricornutum and other diatoms (Siron et al. 1989; Yang et al. 2017; Zhao et al. 2014), fatty acid analysis using GC-MS revealed that palmitic acid (16:0), palmitoleic acid (16:1), and eicosapentaenoic acid (EPA, 20:5 n-3) were the most abundant species detected when using 10% and 30% DMW for cultivation. In addition, the relative abundance of DPA to DHA is consistent with previous reports (Yang et al. 2017). Cultivation using 30% and 60% DMW shifted the fatty acid profile where an increase in DPA content was especially notable compared to F2 and 10% DMW conditions.
This shift to produce more DPA using DMW correlates with the higher initial nitrogen content when more DMW is utilized for cultivation, but the combined data (Supplementary Table 1) suggest that nitrogen content is not the only factor contributing to this effect. Phaeodactylum tricornutum and other diatoms are known to increase SFA and MUFA levels in response to nutrient limitation (Liang et al. 2006; Liang and Mai 2005), and the modification of the initial nitrogen concentration in media has been shown to influence fatty acid composition in P. tricornutum (Qiao et al. 2016). The accumulation of DPA may also be a physiological response to stressors present in the DMW that enhances the resilience in P. tricornutum. As membrane fluidity increases with chain length and degree of unsaturation (Harwood 1988; Jiang and Gao 2004), DPA may contribute to the resilience of P. tricornutum through the relief of turgor pressure, protein recruitment, and nutrient uptake (for review, see Juneja et al. (2013) and Sayanova et al. (2017)). Given that DPA is a commercially valuable omega-3 fatty acid with well-documented human health benefits (Byelashov et al. 2015), increased DPA production using a cheap and abundant alternative nutrient source may have important commercial applications.
Overview of proteomic data comparing P. tricornutum growth on DMW dilutions to growth on synthetic medium
Biochemical and physiological analysis of P. tricornutum cultured in DMW revealed similarities to results observed following nitrogen limitation in this species, with increased lipid contents associated with lower environmental nitrogen concentrations. Therefore, proteomic analysis was performed on P. tricornutum cultures grown on two DMW concentrations that were either nitrogen-limited or nitrogen-equivalent when compared to the nitrogen content of synthetic medium. P. tricornutum was grown on either 10% DMW (nitrogen-limited), 20% DMW (nitrogen-equivalent), or synthetic medium (F2) and sampled for proteomic analysis after 5 days of cultivation (see the “Overview of proteomic data comparing P. tricornutum growth on DMW dilutions to growth on synthetic medium” section and Fig. 3a,b).
Nitrogen metabolism
Proteins involved in nitrogen metabolism were upregulated in the nitrogen-limited 10% DMW condition but not with nitrogen-equivalent 20% DMW compared to F2 (Fig. 4a). P. tricornutum responded to the low environmental nitrogen levels in 10% DMW by increasing its nitrogen uptake and assimilation abilities, as evidenced by the upregulation of a urea transporter (B7FZW5, 10%/F2 + 1.54), nitrate reductase (B7G997, 10%/F2 + 1.35) and glutamate dehydrogenase (B7G3X3, 10%/F2 + 1.30). Upregulation of a xanthine/uracil permease (B7FP05, 10%/F2 + 4.07), purine permease (B7GBV6, 10%/F2 + 2.04), and xanthine dehydrogenase (B7GAV1, 10%/F2 + 1.28) may also provide P. tricornutum with an additional source of nitrogen containing compounds during nitrogen limitation. Cultivation on 10% DMW also led to upregulation of proteins whose functions include the release of ammonia from various macromolecules, including a formidase (B7FYS6, 10%/F2 + 1.89), a putative amidase (B7GE02 + 4.15), two putative cytidine deaminases (B7GCB8, 10%/F2 + 2.08, B7FYE0, 10%/F2 + 2.45), and a protein with hydrolase activity (B7G1X3, 10%/F2 + 2.22) involved in the NAD salvage pathway that catalyzes the release ammonia from nicotinamide. Collectively, upregulation of these proteins supports the conclusion of Longworth and colleagues that P. tricornutum cells remodel their proteome to internally scavenge and recycle nitrogen sources during nitrogen-limited conditions (Longworth et al. 2016).
Iron response
When compared to the F2 control, P. tricornutum grown on either 10% and 20% DMW conditions led to upregulation of multiple proteins associated with an iron limitation response (Fig. 4b) (Allen et al. 2008; Lommer et al. 2012; Morrissey et al. 2015; Morrissey and Bowler 2012). The upregulation of proteins associated with iron limitation suggested that the DMW study was lacking in iron. Subsequent analytical testing of the soluble iron content confirmed the iron-limited nature of the DMW, with the 20% and 10% DMW conditions containing only 3.4% (0.014 mg L-1) and 1.7% (0.007 mg L-1) of the iron found in the F2 medium (0.414 mg L-1), respectively. Diatoms as a group have historically been utilized in environmental monitoring studies (Dixit and Smol 1994) and can survive during periods of low environmental iron availability, with P. tricornutum being particularly tolerant to conditions of low environmental iron (Kustka et al. 2007). The response to low iron levels was especially dramatic in the 20% DMW condition, with four of the top five most upregulated proteins (9–16-fold) identified matching proteins previously linked to an iron starvation response (Allen et al. 2008). These highly induced proteins include iron starvation-induced protein ISIP2A (B7FYL2, 20%/F2 + 4.74), ISIP1 (B7GA90, 20%/F2 + 7.93), ISIP3 (B7G4H8, 20%/F2 + 6.25), and flavodoxin (B7GCM3, 20%/F2 + 6.52). ISIP2A is a cell surface protein whose function in the concentration and uptake of Fe(III) was recently characterized (Morrissey et al. 2015), whereas ISIP1 and ISIP3 are cell surface proteins whose functions during iron limitation remain to be determined. Flavodoxin is an electron-transfer protein that substitutes for ferredoxin in the photosynthetic electron transport during periods of iron limitation.
The expression patterns of iron-responsive proteins in P. tricornutum cultured on 10% DMW conditions suggests that the proteomic response is less robust in 10% DMW compared to 20% DMW condition. Expression of multiple iron response proteins were significantly higher in the 20% DMW condition (Fig. 4b), and the muted iron limitation response observed in the 10% DMW dataset may be in part due to P. tricornutum reacting to the additional stress of low environmental nitrogen that was not present in the 20% DMW condition. Other data from this study support this hypothesis, as P. tricornutum grown in 10% DMW showed a robust response to nitrogen limitation and most likely diverted substantial amounts of energy and resources to synthesize proteins necessary for survival in this environment. These differences are attributed in part to temporal aspects of the iron response and are the result of when sampling occurred for proteomic analysis.
Amino acid metabolism
Differences in the abundance of several proteins involved with amino acid metabolism were observed in P. tricornutum cultures for both 10% and 20% DMW, but meaningful trends in the data were not observed. The upregulation of a vitamin B12-independent methionine synthase (B7G1X4, 10%/F2 + 3.38, 20%/F2 + 2.89) was observed in both 10% DMW and 20% DMW conditions, and this protein is upregulated in cobalamin-limited environments (Bertrand et al. 2012). Additional downregulation of a vitamin B12-dependent methionine synthase (B7GBG7, 10%/F2 -1.21) was observed in the 10% DMW condition. Taken together, differential regulation of these proteins indicates that this DMW source may also be limiting in other micronutrients besides iron.
Photosynthesis
Photosynthesis-related proteins were generally downregulated when comparing the nitrogen-deplete 10% DMW condition to both the nitrogen-equivalent 20% DMW and F2 conditions (Fig. 4c, Supplementary Fig. 1). The most prominent downregulation occurred in pathways controlling biosynthesis of photosynthetic pigments and their precursors. Downregulations of glutamyl-tRNA synthase (B7GCT6, 10%/F2 -1.69, 20%/10% -2.02) and hemL (B7G134, 10%/F2 -1.20) indicate a reduction in the biosynthesis of tetrapyrroles, compounds with important functions as cofactors in chlorophyll pigments, as well as heme-containing proteins. Decreased expressions of proteins hemE (B7GB77, 10%/F2 -1.74), PPO (B7GDU9, 10%/F2 -2.00, 10%/20% -1.67), CHLD (B7FTA2, 10%/F2 -1.75, 10%/20% -1.43), and DVR (B7GC47, 10%/F2 -1.75) indicate that chlorophyll biosynthesis was also downregulated. Biosynthesis of carotenoids was also downregulated, as evidenced by the downregulation of proteins involved in biosynthesis of isoprenoids (HDS, B7FV10 10%/F2 -1.26; GGPS, B7FU89, 10%/F2 -1.74, 10%/20%, -1.59) and carotenoids (PDS1, B7FZL9, 10%/F2 -1.48; ZDS, B7FPC4, 10%/F2 -1.18; CRTISO5, B7FQF7, 10%/20% -1.67). No evidence of downregulation of chlorophyll and carotenoid biosynthesis was observed when comparing the 20% DMW condition to the F2 control.
Multiple light-harvesting complex (LHCs) fucoxanthin-chlorophyll a/c binding proteins (FCPs) were observed upregulated in the 10% and 20% DMW conditions compared to the F2 control (Fig. 4c). Unlike what has been observed in other proteomic experiments investigating nitrogen limitation in P. tricornutum, cultivation in 10% DMW culture did not lead to a general decrease in the expression of chlorophyll binding light-harvesting protein, and the pattern of regulation in these proteins resembles the 20% DMW condition. Iron deficiency has been shown to cause a rearrangement of the photosynthetic apparatus in diatoms (Allen et al. 2008) and may influence light-harvesting proteins in an effort to increase photoprotection mechanisms. Indeed, LHCX2 (B7FR60, 20%/F2 + 2.85, 10%/F2 + 2.51), an important antenna protein involved in nonphotochemical quenching (NPQ) in P. tricornutum (Lepetit et al. 2017; Nymark et al. 2009), was upregulated in both 10% and 20% DMW conditions. Flavodoxin (B7GCM3, 20%/F2 + 6.52, 10%/F2 + 6.70), as described above, was upregulated in both 10% and 20% DMW conditions when compared to the F2 control and was the only differentially regulated protein specific to the photosynthetic apparatus.
Rubisco activase (A0T0M5, 10%/F2 -1.14) and the carbonic anhydrase isoform CA4 (B7FNU0, 10%/F2 -1.92) were downregulated in the 10% DMW condition, suggesting the decreased role of photosynthetic carbon fixation during periods of nitrogen limitation. A candidate bicarbonate transporter part of the SLC4 family (B7S437, 20%/F2 -2.65, 10%/F2 -2.47) was also downregulated in both 10% and 20% DMW conditions. Finally, FBA5C (B7GE67, 20%/F2 + 4.66, 10%/F2 + 3.12), a plastidic fructose bisphosphate aldolase isoform, was upregulated in both 10% and 20% DMW conditions. It has been hypothesized that during times of low carbon assimilation, a condition observed during iron limitation in P. tricornutum (Allen et al. 2008), the reaction FBAC5 catalyzes may run in the reverse direction, consequently supplying carbon skeletons for anabolic reactions (Allen et al. 2012).
Protein metabolism
After 5 days of cultivation on 10% and 20% DMW, differential regulation of multiple proteins involved in protein synthesis and degradation was observed (Fig. 5). Compared to the F2 control, P. tricornutum cultured on 10% DMW led to downregulation of 19 ribosomal structural proteins, indicating an overall decrease in protein synthesis. Four proteins with annotated proteolytic functions were upregulated (all 10%/F2: B7GDQ0, + 1.60; B7FSD0, + 3.25; B7FUY6, + 1.25; B7G5X7, + 2.36), while one protease (B5Y4I2, 10%/F2 -1.89) and a ubiquitin extension protein (B7G5H7, 10%/F2 -2.26) were downregulated. P. tricornutum grown in 20% DMW also exhibited mixed regulation of proteins involved in protein degradation with two proteases upregulated (all 20%/F2: B7FSD0, + 3.07; B7FVG8, + 1.73) and one protease downregulated (B5Y4I2, 20%/F2 -2.85). However, in contrast to the 10% DMW condition, the downregulation of ribosomal structural proteins was not observed, and five ribosomal structural proteins (all 20%/F2: A0T0J5, + 1.45; B7FP95, + 2.85; A0T0C7, + 1.80; A0T0I4, + 1.91; B7FQC3, + 2.22) were upregulated when compared to the F2 control.
MapMan-based visualization of significantly (p < 0.05, log2FC ± 1) regulated proteins involved in protein metabolism in a F2 compared to 10% DMW and b F2 compared to 20% DMW. Proteins are represented as individual blocks and colored blue or red if upregulated or downregulated in abundance, respectively
Proteins involved in regulating protein synthesis, including those involved in translation initiation (B7G3T4, 20%/F2 + 1.58, 10%/F2 + 1.56; B7FV44, 20%/F2 2.23, 10%/F2 2.91) and elongation (B7G3C4, 20%/F2 + 1.53, 10%/F2 + 1.44), were upregulated in both 10% and 20% DMW conditions, with an additional protein with functions in translation initiation (B7G5G1, 10%/F2 + 1.78) upregulated in the 10% DMW condition only. Taken together, this regulation pattern supports the conclusion that P. tricornutum grown in 10% DMW decreases protein synthesis while simultaneously enhancing targeted protein degradation. This response may function as part of a mechanism that reduces protein synthesis as a general sink for nitrogen while also increasing nitrogen availability for the biosynthesis of specific proteins during periods of low environmental nitrogen availability. Organisms exert a tight control on protein synthesis and degradation in response to fluctuating environmental conditions, and the mixed regulation of proteases combined with the upregulation of ribosomal structural proteins and of proteins with translation initiation and elongation factor activity suggest the importance of controlling protein synthesis during cultivation on this DMW source.
Central carbon metabolism and oxidative phosphorylation
A broad upregulation of proteins involved in central carbon metabolism and oxidative phosphorylation were observed in P. tricornutum cultivated on 10% DMW, including enzymes associated with glycolysis (B7GC94, 10%/F2 + 1.27; B7G8B7, 10%/F2 + 1.73), the TCA cycle (all 10%/F2: B7FZD7, + 1.39; B7G228, + 1.27; B7FWX5, + 2.35), and oxidative phosphorylation (all 10%/F2 B7GBT9, + 2.14; B7FXG3, + 1.19; B7G964, + 2.11; B7G360, + 1.61) (Fig. 4d). Upregulation of dihydroorotate dehydrogenase (B7G8S8, + 1.91) during cultivation on 10% DMW was also observed, which functions in pyrimidine biosynthesis and energy generation at the mitochondrial membrane. The additional upregulation of two putative mitochondrial phosphate transporters observed following cultivation on 10% DMW (B7G6E4, 10%/F2 + 1.27) or 10% and 20% DMW conditions (B7G5H6, 20%/F2 + 1.86, 10%/F2 + 1.67), respectively, and may function to supply inorganic phosphate to the mitochondria for ATP biosynthesis.
Interestingly, cultivation on 10% and 20% DMW dilutions resulted in an upregulation of two short chain dehydrogenase/reductases (SDR) (B7G0Q2, 20%/F2 + 1.99, 10%/F2 + 1.54; B7GBR2, 20%/F2 + 2.11, 10%/F2 + 2.61) and an aldo-keto reductase (AKR) (B7GDN9, 20%/F2 + 1.78, 10%/F2 + 1.84) when compared to the F2 control. Although SDR and AKR protein families are very diverse in their structure and functions, both families contain NAD+/NADP+ binding domains and can participate in oxidoreductase reactions that reduce aldehydes and ketones to primary and secondary alcohols, respectively (Penning 2015). The downregulation of glyoxalase II (B7GDI1, 20%/F2 -2.20) in the 20% DMW condition could mean that the observed upregulation of AKR and SDR proteins with unknown specific functions may be involved in the detoxification of methylglyoxal (Grant et al. 2003; Yamauchi et al. 2011), a toxic byproduct of carbon metabolism. SDR and AKR protein families also have known roles in redox sensing (Kavanagh et al. 2008) and stress tolerance (Kanayama et al. 2014), and three additional alcohol dehydrogenases (all 10%/F2: B7GEB0, + 1.44; B7GEA6, + 1.44; B7S4B2, + 1.35) of unknown specificity that were upregulated following 10% DMW cultivation may have been upregulated in response to low levels of environmental nitrogen.
Redox/stress
Analysis of the proteome suggests that P. tricornutum responds to DMW conditions by increasing the abilities to regulate oxidative stress levels and maintain redox homeostasis (Fig. 4e). The increased expression of glutathione reductase (GSR_1, B7G9Q1, 10%/F2 + 2.01) in 10% DMW and glutathione synthase (B5Y5J7, 20%/F2 + 1.78) in 20% DMW suggests that P. tricornutum attempts to increase or replenish reduced glutathione (GSH) levels in response to DMW conditions. Maintaining high levels of reduced glutathione pools have been shown to increase abiotic stress tolerance in Arabidopsis (Cheng et al. 2015) and may enhance the ROS detoxification ability of P. tricornutum. Thioredoxin h (B7G7L6, 20%/F2 + 2.10, 10%/F2 + 2.18) was upregulated in both 10% and 20% DMW conditions. H-type thioredoxins have known functions in oxidative stress management (Serrato and Cejudo 2003) and may be involved in the redox regulation of glutathione reductases as seen in Triticum aestivum (Gelhaye et al. 2003). B7G7L6 shares a 40.4% identity with Trxh1 in Oryza sativa, which is induced during abiotic stress, including an elucidated function in managing reactive oxygen species balance in the rice apoplast during salt stress (Zhang and Guo 2012). Increased expression of a Mpv17/PMP-like protein (B7G8P1, 20%/F2 + 1.46) targeting to the peroxisomal membrane provides further evidence of ROS management in the 20% DMW condition. Nitrogen starvation produces oxidative microenvironments in P. tricornutum (Rosenwasser et al. 2014), and additional upregulation of a peroxiredoxin (B7GCV6, 10%/F2 + 3.43), an NADP+ transhydrogenase (B5Y5A6, 10%/F2 + 1.79), and putative glutathione-s-transferase (B5Y512, 10%/F2 + 1.44) in the 10% DMW condition may be required to combat oxidative stress during nitrogen limitation.
Cellular signaling and gene expression
P. tricornutum grown on both 10% and 20% DMW for 5 days exhibited alterations in expression profiles for multiple proteins involved in cellular signaling (Fig. 4f). Compared to the F2 control, hCDK3 (B7FUX2, 10%/F2 + 145) was upregulated in 10% DMW, and CDKA2 (B7GDW6, 20%/F2 -1.41) was downregulated in 20% DMW, suggesting altered regulation of the cell cycle during cultivation on DMW. The upregulations of two calcium-dependent kinases (B7GBS5, 20%/F2 + 1.66, 10%/F2 + 2.86; B7G8H4, 20%/F2 + 2.82, 10%/F2 + 3.33) in both 10% and 20% DMW, along with the upregulation of phospholipase C (B7S3T2, 10%/F2 + 1.78) and copine 1 (B7FSV2, 10%/F2 + 1.30) in 10% DMW, suggest that calcium signaling plays an important role in acclimatizing to culture on this DMW source. Culture on both DMW conditions also downregulated a P-type ATPase (B7GCD8 20%/F2 -1.84, 10%/F2 -1.54) identified as a calcium ion carrier, providing further evidence of modulated calcium signaling pathways. It must be noted, however, that P. tricornutum utilizes calcium signaling as part of its iron limitation response (Allen et al. 2008; Falciatore et al. 2000), and given the iron-limited composition of the DMW used in this study, further studies must be performed to confirm and elucidate the importance of calcium-based signaling during acclimatization to culture on DMW sources.
Three proteins with roles that function as molecular chaperones were downregulated in the 20% DMW condition, including a DnaJ (hsp40) homolog (B7G4P7, 20%/F2 -4.17), an hsp90 homolog (B7GES3, 20%/F2 -4.52), and the molecular chaperone cdc37 (B7S3L1, 20%/F2 -1.60). These proteins are components within a multi-protein complex that function to chaperone various targets, including protein kinases (Arlander et al. 2006). The functional consequence of their downregulation in the 20% DMW condition is unknown but signify further evidence of alterations in signal transduction.
Phaeodactylum tricornutum grown on DMW exhibited differences in proteins involved in transcriptional regulation when compared to culture on synthetic medium. Two transcription factors, one identified as containing an HMG box domain (B7FQD6, 20%/F2, -4.25) and another identified Myb1R2 (B7G7Q5, 20%/F2 -2.32) (Rayko et al. 2010), were downregulated in 20% DMW. An additional protein identified as splicing factor 3B subunit 3 (B7FP73, 20%/F2 -1.97), which functions in pre-mRNA splicing, was also downregulated in the 20% DMW. The CO2 responsive bZIP11 (B7G9T9, 10%/F2 + 1.34) transcription factor was upregulated in 10% DMW, while CSF4 (B7G5B5, 20%/F2 + 2.62, 10%/F2 + 2.77), a DNA-binding protein containing a cold shock domain, was upregulated in both 10% and 20% DMW. Although this study cannot clearly implicate the functional consequences that result from altering transcriptional regulation and cellular signaling pathways, it may contribute to the proteomic re-organization observed in P. tricornutum in response to DMW cultivation.
Membrane transport and trafficking
Proteomic analysis after 5 days of growth on DMW suggests that P. tricornutum responds to DMW conditions by controlling small molecule and protein transport throughout various cellular compartments. Upregulation of small GTPases belonging to the Ras superfamily of proteins were observed in both 10% and 20% conditions, including proteins identified as RabX1 (B7S3S7, 20%/F2 + 1.92, 10%/F2 + 2.29) and Arf1 (B7FR50, 20%/F2 + 3.97, 10%/F2 + 4.32), with the Rab GTPase Sec4 (B7G9L9, 10%/F2 + 1.79) observed upregulated in the 10% DMW condition only (Supplementary Fig. 2). Both RabX1, which shares a 78.9% sequence identify with the Ras-related protein Rab-8A in Fragilariopsis cylindrus, and Arf1 are involved in endocytosis. Additional support of increased endocytic pathways was observed in the 10% DMW condition, where upregulation of adaptor protein complex components AP1/2beta (B7S4C6, 10%/F2 + 1.23) and AP2M1 (B7FSB1, 10%/F2 + 1.48) was observed (Supplementary Fig. 3). The increased expression of a V-type H+ transporting ATPase (B7G360, 10%/F2 + 1.61) in 10% DMW suggests an increased role for phagocytosis in this condition and, taken together, could provide P. tricornutum grown on the DMW an additional scavenging mechanism to acquire limiting nutrients such as iron and nitrogen from the outside environment.
Protein trafficking was generally upregulated during DMW culture. The nuclear transport factor NFT2 (B7G8V7, 20%/F2 + 1.57, 10%/F2 + 2.21) was upregulated in both conditions, and the Ran-GTPase importin-7 (B7FVE6, 10%/F2 + 1.21) was upregulated in 10% DMW. Nuclear export by exportin-1 (B7GEG3, 20%/F2 -1.68) was downregulated in 20% DMW. The alpha subunit of sec61 (B7GD39, 20%/F2 + 1.79, 10%/F2 + 1.35), sec31 (B7G255, 20%/F2 + 1.63, 10%/F2 + 2.20), and a protein identified as part of the SRP-independent protein family (B7FUG7, 20%/F2 + 1.44, 10%/F2 + 1.57) were upregulated in 10% and 20% DMW conditions, suggesting an increase in protein translocation through the endoplasmic reticulum (Supplementary Fig. 2), and provide support for the importance of controlling protein synthesis during DMW culturing.
Cultivation on DMW increased the expression of a MFS superfamily transport protein (B7FTZ4, 20%/F2 + 1.46, 10%/F2 + 1.60) that putatively functions in the transport of molybdate ions across membranes. In its biologically active form, molybdenum acts as a cofactor in specific enzymes, including purine catabolism such as xanthine dehydrogenase (B7GAV1, 10%/F2 + 1.28) and nitrogen assimilation nitrate reductase (B7G997, 10%/F2 + 1.35), both of which were upregulated in the 10% DMW condition. Two additional putative phosphate transporters (B7GBF0, 20%/F2 -3.90 and B7G4H1, 20%/F2 -2.48) were also downregulated in 20% DMW.
Lipid metabolism
Phaeodactylum tricornutum cultivated on either 10% or 20% DMW upregulated an acyl carrier protein (ACP) (B7FRX6, 20%/F2 + 2.50, 10%/F2 + 2.46) and downregulated a stearoyl-ACP desaturase (B7FQK1, 20%/F2 -2.01, 10%/F2 -1.92). ACPs perform important functions as cofactors in fatty acid biosynthesis, and overexpressions of specific ACP isoforms have been demonstrated to alter FA composition (Branen et al. 2001) and modulate salt stress tolerance in Arabidopsis (Huang et al. 2017). Stearoyl-ACP desaturase requires iron as a cofactor, and iron limitation in this medium source may explain the observed downregulation. A decrease in FA desaturase is expected to alter the fatty acid profile towards increased saturation, and in C. reinhardtii, iron starvation downregulates expression of multiple FA desaturase genes, increasing the saturated to unsaturated FA ratio while altering the FA composition of membrane lipids (Urzica et al. 2013). Interestingly, a recent study where proteomic analysis was performed on of P. tricornutum grown on iron-limited or iron-starved F2 medium did not cause downregulation of FA desaturases (Zhao et al. 2018), highlighting microalgae species-specific differences in responding to iron limitation. Indeed, the FA composition of P. tricornutum cultured on DMW did not increase FA saturation when compared to culture on F2, and FA unsaturation actually increased in all DMW dilution used besides the nitrogen-limited 10% DMW condition (Table 2).
Conclusions
This study demonstrates the utility of dairy manure wastewater as a media source for P. tricornutum cultivation. The fatty acid composition, biomass yields, and lipid yields vary based on the dilution of DMW in seawater (from 10 to 60%), which correlates to starting nitrogen content. Alterations in the proteome identified across cultivation conditions demonstrate that P. tricornutum responds to DMW cultivation by altering multiple biological pathways. This study demonstrates that proteomic analysis can contribute to wastewater characterization, e.g., to identify the low iron content of DMW. The demonstration of successful cultivation and significant lipid accumulation of P. tricornutum on DMW supports the viability of using low-cost waste nutrients for microalgae biomass and biofuel production, as well as the partial remediation of wastewaters during microalgae cultivation. Overall, this study contributes to greater insight of proteome reorganization of microalgae during wastewater cultivation.
Data Availability
All data for this manuscript is available in the supplementary information or by request to the corresponding author.
References
Allen AE, LaRoche J, Maheswari U, Lommer M, Schauer N, Lopez PJ, Finazzi G, Fernie AR, Bowler C (2008) Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc Natl Acad Sci 105:10438–10443
Allen AE, Moustafa A, Montsant A, Eckert A, Kroth PG, Bowler C (2012) Evolution and functional diversification of fructose bisphosphate aldolase genes in photosynthetic marine diatoms. Mol Biol Evol 29:367–379
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115–D119
Arlander SJH, Felts SJ, Wagner JM, Stensgard B, Toft DO, Karnitz LM (2006) Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J Biol Chem 281:2989–2998
Bertrand EM, Allen AE, Dupont CL, Norden-Krichmar TM, Bai J, Valas RE, Saito MA (2012) Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc Natl Acad Sci 109:E1762–E1771
Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, de Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, la Roche J, Lindquist E, Lommer M, Martin–Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, Montsant A, Secq MPO–L, Napoli C, Obornik M, Parker MS, Petit JL, Porcel BM, Poulsen N, Robison M, Rychlewski L, Rynearson TA, Schmutz J, Shapiro H, Siaut M, Stanley M, Sussman MR, Taylor AR, Vardi A, von Dassow P, Vyverman W, Willis A, Wyrwicz LS, Rokhsar DS, Weissenbach J, Armbrust EV, Green BR, van de Peer Y, Grigoriev IV (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Branen JK, Chiou T-J, Engeseth NJ (2001) Overexpression of acyl carrier protein-1 alters fatty acid composition of leaf tissue in Arabidopsis. Plant Physiol 127:222–229
Burch AR, Franz AK (2016) Combined nitrogen limitation and hydrogen peroxide treatment enhances neutral lipid accumulation in the marine diatom Phaeodactylum tricornutum. Bioresour Technol 219:559–565
Byelashov OA, Sinclair AJ, Kaur G (2015) Dietary sources, current intakes, and nutritional role of omega-3 docosapentaenoic acid. Lipid Technol 27:79–82
Chen W, Zhang C, Song L, Sommerfeld M, Hu Q (2009) A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J Microbiol Methods 77:41–47
Cheng M, Ko K, Chang W, Kuo W, Chen G, Lin T (2015) Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J 83:926–939
Chokshi K, Pancha I, Ghosh A, Mishra S (2016) Microalgal biomass generation by phycoremediation of dairy industry wastewater: an integrated approach towards sustainable biofuel production. Bioresour Technol 221:455–460
Coppens J, Decostere B, Van Hulle S, Nopens I, Vlaeminck SE, De Gelder L, Boon N (2014) Kinetic exploration of nitrate-accumulating microalgae for nutrient recovery. Appl Microbiol Biotechnol 98:8377–8387
Demirer GN, Chen S (2005) Two-phase anaerobic digestion of unscreened dairy manure. Process Biochem 40:3542–3549
Dixit SS, Smol JP (1994) Diatoms as indicators in the environmental monitoring and assessment program-surface waters (EMAP-SW). Environ Monit Assess 31:275–307
Fabris M, Matthijs M, Rombauts S, Vyverman W, Goossens A, Baart GJE (2012) The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner–Doudoroff glycolytic pathway. Plant J 70:1004–1014
Falciatore A, d’Alcalà MR, Croot P, Bowler C (2000) Perception of environmental signals by a marine diatom. Science 288:2363–2366
Fargione J, Hill J, Tilman D, Polasky S, Hawthorne P (2008) Land clearing and the biofuel carbon debt. Science 319:1235–1238
Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226:497–509
Franz AK, Danielewicz MA, Wong DM, Anderson LA, Boothe JR (2013) Phenotypic screening with oleaginous microalgae reveals modulators of lipid productivity. ACS Chem Biol 8:1053–1062
Gelhaye E, Rouhier N, Jacquot J-P (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett 555:443–448
Grant AW, Steel G, Waugh H, Ellis EM (2003) A novel aldo-keto reductase from Escherichia coli can increase resistance to methylglyoxal toxicity. FEMS Microbiol Lett 218:93–99
Hannon M, Gimpel J, Tran M, Rasala B, Mayfield S (2010) Biofuels from algae: challenges and potential. Biofuels 1:763–784
Harwood JL (1988). Fatty acid metabolism. Ann Rev Plant Physiol Plant Mol Biol 39:101–138
Higgins BT, Thornton-Dunwoody A, Labavitch JM, VanderGheynst JS (2014) Microplate assay for quantitation of neutral lipids in extracts from microalgae. Anal Biochem 465:81–89
Ho S-H, Nakanishi A, Kato Y, Yamasaki H, Chang J-S, Misawa N, Hirose Y, Minagawa J, Hasunuma T, Kondo A (2017) Dynamic metabolic profiling together with transcription analysis reveals salinity-induced starch-to-lipid biosynthesis in alga Chlamydomonas sp. JSC4. Sci Rep 7:45471
Hörtensteiner S, Kräutler B (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta-Bioenergetics 1807:977–988
Huang J, Xue C, Wang H, Wang L, Schmidt W, Shen R, Lan P (2017) Genes of acyl carrier protein family show different expression profiles and overexpression of acyl carrier protein 5 modulates fatty acid composition and enhances salt stress tolerance in Arabidopsis. Front Plant Sci 8:987
Ichihara K, Fukubayashi Y (2010) Preparation of fatty acid methyl esters for gas-liquid chromatography. J Lipid Res 51:635–640
Jiang H, Gao K (2004) Effects of lowering termperature during culture on the production of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum (Bacillariophyceae). J Phycol 40:651–654
Jiang L, Luo S, Fan X, Yang Z, Guo R (2011) Biomass and lipid production of marine microalgae using municipal wastewater and high concentration of CO2. Appl Energy 88:3336–3341
Joachimiak MP, Weisman JL, May BCH (2006) JColorGrid: software for the visualization of biological measurements. BMC Bioinform 7:225
Juneja A, Ceballos R, Murthy G (2013) Effects of environmental factors and nutrient availability on the biochemical composition of algae for biofuels production: a review. Energies 6:4607–4638
Kanayama Y, Mizutani R, Yaguchi S, Hojo A, Ikeda H, Nishiyama M, Kanahama K (2014) Characterization of an uncharacterized aldo-keto reductase gene from peach and its role in abiotic stress tolerance. Phytochemistry 104:30–36
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114
Kavanagh KL, Jörnvall H, Persson B, Oppermann U (2008) Medium-and short-chain dehydrogenase/reductase gene and protein families. Cell Mol Life Sci 65:3895–3906
Kustka AB, Allen AE, Morel FMM (2007) Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J Phycol 43:715–729
Lepetit B, Gélin G, Lepetit M, Sturm S, Vugrinec S, Rogato A, Kroth PG, Falciatore A, Lavaud J (2017) The diatom Phaeodactylum tricornutum adjusts nonphotochemical fluorescence quenching capacity in response to dynamic light via fine-tuned Lhcx and xanthophyll cycle pigment synthesis. New Phytol 214:205–218
Liang Y, Beardall J, Heraud P (2006) Changes in growth, chlorophyll fluorescence and fatty acid composition with culture age in batch cultures of Phaeodactylum tricornutum and Chaetoceros muelleri (Bacillariophyceae). Bot Mar 49:165–173
Liang Y, Mai K (2005) Effect of growth phase on the fatty acid compositions of four species of marine diatoms. J Ocean Univ China 4:157–162
Libralato G, Gentile E, Volpi Ghirardini A (2016) Wastewater effects on Phaeodactylum tricornutum (Bohlin): setting up a classification system. Ecol Indic 60:31–37
Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B (2014) Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ 37:1250–1258
Lommer M, Specht M, Roy A-S, Kraemer L, Andreson R, Gutowska MA, Wolf J, Bergner SV, Schilhabel MB, Klostermeier UC, Beiko RG, Rosenstiel P, Hippler M, LaRoche J (2012) Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol 13:R66–R66
Longworth J, Wu D, Huete-Ortega M, Wright PC, Vaidyanathan S (2016) Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res 18:213–224
Moreno-Caselles J, Moral R, Perez-Murcia M, Perez-Espinosa A, Rufete B (2002) Nutrient value of animal manures in front of environmental hazards. Commun Soil Sci Plant Anal 33:3023–3032
Morrissey J, Bowler C (2012) Iron utilization in marine cyanobacteria and eukaryotic algae. Front Microbiol 3:43
Morrissey J, Sutak R, Paz-Yepes J, Tanaka A, Moustafa A, Veluchamy A, Thomas Y, Botebol H, Bouget F-Y, McQuaid JB, Tirichine L, Allen AE, Lesuisse E, Bowler C (2015) A novel protein, ubiquitous in marine phytoplankton, concentrates iron at the cell surface and facilitates uptake. Curr Biol 25:364–371
Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I (2013) The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res 42:D26–D31
Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM (2009) An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS One 4:e7743
Park J, Wang H, Gargouri M, Deshpande RR, Skepper JN, Holguin FO, Juergens MT, Shachar-Hill Y, Hicks LM, Gang DR (2015) The response of Chlamydomonas reinhardtii to nitrogen deprivation: a systems biology analysis. Plant J 81:611–624
Patel AK, Huang EL, Low-Décarie E, Lefsrud MG (2015) Comparative shotgun proteomic analysis of wastewater-cultured microalgae: nitrogen sensing and carbon fixation for growth and nutrient removal in Chlamydomonas reinhardtii. J Proteome Res 14:3051–3067
Penning TM (2015) The aldo-keto reductases (AKRs): overview. Chem Biol Interact 234:236–246
Qiao H, Cong C, Sun C, Li B, Wang J, Zhang L (2016) Effect of culture conditions on growth, fatty acid composition and DHA/EPA ratio of Phaeodactylum tricornutum. Aquaculture 452:311–317
Rawat I, Gupta SK, Shriwastav A, Singh P, Kumari S, Bux F (2016) Microalgae applications in wastewater treatment. In: Bux F, Chisti Y (eds) Algae Biotechnology: Products and processes. Springer, Cham, pp 249–268
Rayko E, Maumus F, Maheswari U, Jabbari K, Bowler C (2010) Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol 188:52–66
Ritchie RJ (2006) Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth Res 89:27–41
Rosenwasser S, van Creveld SG, Schatz D, Malitsky S, Tzfadia O, Aharoni A, Levin Y, Gabashvili A, Feldmesser E, Vardi A (2014) Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc Natl Acad Sci 111:2740–2745
Sayanova O, Mimouni V, Ulmann L, Morant-Manceau A, Pasquet V, Schoefs B, Napier JA (2017) Modulation of lipid biosynthesis by stress in diatoms. Phil Trans Roy Soc B 372:20160407
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Serrato AJ, Cejudo FJ (2003) Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta 217:392–399
Simon DF, Domingos RF, Hauser C, Hutchins CM, Zerges W, Wilkinson KJ (2013) Transcriptome sequencing (RNA-seq) analysis of the effects of metal nanoparticle exposure on the transcriptome of Chlamydomonas reinhardtii. Appl Environ Microbiol 79:4774–4785
Siron R, Giusti G, Berland B (1989) Changes in the fatty acid composition of Phaeodactylum tricornutum and Dunaliella tertiolecta during growth and under phosphorus deficiency. Mar Ecol Prog Ser 55:95–100
Smith SA, Hughes E, Coats ER, Brinkman CK, McDonald AG, Harper JR, Feris K, Newby D (2016) Toward sustainable dairy waste utilization: enhanced VFA and biogas synthesis via upcycling algal biomass cultured on waste effluent. J Chem Technol Biotechnol 91:113–121
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Urzica EI, Vieler A, Hong-Hermesdorf A, Page MD, Casero D, Gallaher SD, Kropat J, Pellegrini M, Benning C, Merchant SS (2013) Remodeling of membrane lipids in iron-starved Chlamydomonas. J Biol Chem 288:30246–30258
Valenzuela J, Carlson RP, Gerlach R, Cooksey K, Peyton BM, Bothner B, Fields MW (2013) Nutrient resupplementation arrests bio-oil accumulation in Phaeodactylum tricornutum. Appl Microbiol Biotechnol 97:7049–7059
Yamauchi Y, Hasegawa A, Taninaka A, Mizutani M, Sugimoto Y (2011) NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J Biol Chem 286:6999–7009
Yang Y-H, Du L, Hosokawa M, Miyashita K, Kokubun Y, Arai H, Taroda H (2017) fatty acid and lipid class composition of the microalga Phaeodactylum tricornutum. J Oleo Sci 66:363–368
Yang Z-K, Ma Y-H, Zheng J-W, Yang W-D, Liu J-S, Li H-Y (2014) Proteomics to reveal metabolic network shifts towards lipid accumulation following nitrogen deprivation in the diatom Phaeodactylum tricornutum. J Appl Phycol 26:73–82
Yodsuwan N, Sawayama S, Sirisansaneeyakul S (2017) Effect of nitrogen concentration on growth, lipid production and fatty acid profiles of the marine diatom Phaeodactylum tricornutum. Agric Nat Resour 51:190–197
Zhang C-J, Guo Y (2012) OsTRXh1 regulates the redox state of the apoplast and influences stress responses in rice. Plant Signal Behav 7:440–442
Zhao P, Gu W, Huang A, Wu S, Liu C, Huan L, Gao S, Xie X, Wang G (2018) Effect of iron on the growth of Phaeodactylum tricornutum via photosynthesis. J Phycol 54:34–43
Zhao P, Gu W, Wu S, Huang A, He L, Xie X, Gao S, Zhang B, Niu J, Peng Lin A, Wang G (2014) Silicon enhances the growth of Phaeodactylum tricornutum Bohlin under green light and low temperature. Sci Rep 4:3958
Funding
This work is supported by the National Science Foundation - Chemical, Bioengineering, Environmental, and Transport Systems (NSF-CBET) Award under Grant No. 1512250.
Author information
Authors and Affiliations
Contributions
ARB contributed to the conception, experimental design, data acquisition, analysis, and drafting of the article. CWY contributed to the experimental design, data acquisition, and analysis of the FAME data presented in the article and also contributed to the drafting and critical revision of this article. PP collected and contributed the flushed dairy manure wastewater utilized in the study and provided critical review of the manuscript. MS contributed to proteomic data acquisition and troubleshooting associated with processing algae biomass for proteomic analysis. BP contributed to the bioinformatic analysis of proteomic data presented in the article and also contributed to the drafting and critical revision of this article. AKF contributed to the conception, design, supervision, drafting, and critical revision of this article. All authors give their final approval of the submitted manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 198 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Burch, A.R., Yothers, C.W., Salemi, M.R. et al. Quantitative label-free proteomics and biochemical analysis of Phaeodactylum tricornutum cultivation on dairy manure wastewater. J Appl Phycol 33, 2105–2121 (2021). https://doi.org/10.1007/s10811-021-02483-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-021-02483-3